Alternative Transposition Generates New Chimeric Genes and Segmental Duplications at the Maize p1 Locus
- PMID: 26434719
- PMCID: PMC4649661
- DOI: 10.1534/genetics.115.178210
Alternative Transposition Generates New Chimeric Genes and Segmental Duplications at the Maize p1 Locus
Abstract
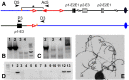
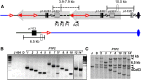
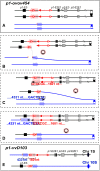
The maize Ac/Ds transposon family was the first transposable element system identified and characterized by Barbara McClintock. Ac/Ds transposons belong to the hAT family of class II DNA transposons. We and others have shown that Ac/Ds elements can undergo a process of alternative transposition in which the Ac/Ds transposase acts on the termini of two separate, nearby transposons. Because these termini are present in different elements, alternative transposition can generate a variety of genome alterations such as inversions, duplications, deletions, and translocations. Moreover, Ac/Ds elements transpose preferentially into genic regions, suggesting that structural changes arising from alternative transposition may potentially generate chimeric genes at the rearrangement breakpoints. Here we identified and characterized 11 independent cases of gene fusion induced by Ac alternative transposition. In each case, a functional chimeric gene was created by fusion of two linked, paralogous genes; moreover, each event was associated with duplication of the ∼70-kb segment located between the two paralogs. An extant gene in the maize B73 genome that contains an internal duplication apparently generated by an alternative transposition event was also identified. Our study demonstrates that alternative transposition-induced duplications may be a source for spontaneous creation of diverse genome structures and novel genes in maize.
Keywords: Ac/Ds alternative transposition; chimeric gene; exon shuffling; segmental duplication; translocation.
Copyright © 2015 by the Genetics Society of America.
Figures




References
-
- Bennetzen J. L., 2005. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15: 621–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

