Review of the Structural and Dynamic Mechanisms of PPARγ Partial Agonism
- PMID: 26435709
- PMCID: PMC4578752
- DOI: 10.1155/2015/816856
Review of the Structural and Dynamic Mechanisms of PPARγ Partial Agonism
Abstract
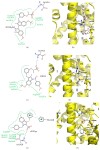
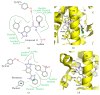
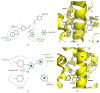
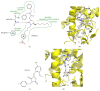
PPARγ (peroxisome proliferator activated receptor γ) is a ligand activated transcription factor of the nuclear receptor superfamily that controls the expression of a variety of genes involved in fatty acid metabolism, adipogenesis, and insulin sensitivity. While endogenous ligands of PPARγ include fatty acids and eicosanoids, synthetic full agonists of the receptor, including members of the thiazolidinedione (TZD) class, have been widely prescribed for the treatment of type II diabetes mellitus (T2DM). Unfortunately, the use of full agonists has been hampered by harsh side effects with some removed from the market in many countries. In contrast, partial agonists of PPARγ have been shown to retain favourable insulin sensitizing effects while exhibiting little to no side effects and thus represent a new potential class of therapeutics for the treatment of T2DM. Partial agonists have been found to not only display differences in transcriptional and cellular outcomes, but also act through distinct structural and dynamic mechanisms within the ligand binding cavity compared to full agonists.
Figures



References
-
- Braissant O., Foufelle F., Scotto C., Dauça M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137(1):354–366. - PubMed
-
- Gearing K. L., Göttlicher M., Teboul M., Widmark E., Gustafsson J.-Å. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(4):1440–1444. doi: 10.1073/pnas.90.4.1440. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

