Standards not that standard
- PMID: 26435739
- PMCID: PMC4591577
- DOI: 10.1186/s13036-015-0017-9
Standards not that standard
Abstract
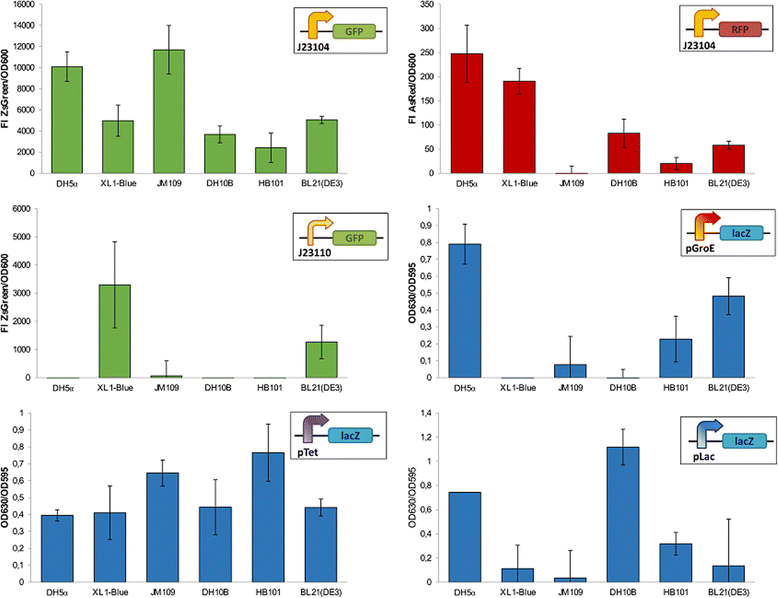
There is a general assent on the key role of standards in Synthetic Biology. In two consecutive letters to this journal, suggestions on the assembly methods for the Registry of standard biological parts have been described. We fully agree with those authors on the need of a more flexible building strategy and we highlight in the present work two major functional challenges standardization efforts have to deal with: the need of both universal and orthogonal behaviors. We provide experimental data that clearly indicate that such engineering requirements should not be taken for granted in Synthetic Biology.
Keywords: Biobrick parts; Orthogonality; Standardization; Synthetic biology.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

