Antisense Oligonucleotide-Mediated Transcript Knockdown in Zebrafish
- PMID: 26436892
- PMCID: PMC4593562
- DOI: 10.1371/journal.pone.0139504
Antisense Oligonucleotide-Mediated Transcript Knockdown in Zebrafish
Abstract
Antisense oligonucleotides (ASOs) are synthetic, single-strand RNA-DNA hybrids that induce catalytic degradation of complementary cellular RNAs via RNase H. ASOs are widely used as gene knockdown reagents in tissue culture and in Xenopus and mouse model systems. To test their effectiveness in zebrafish, we targeted 20 developmental genes and compared the morphological changes with mutant and morpholino (MO)-induced phenotypes. ASO-mediated transcript knockdown reproduced the published loss-of-function phenotypes for oep, chordin, dnd, ctnnb2, bmp7a, alk8, smad2 and smad5 in a dosage-sensitive manner. ASOs knocked down both maternal and zygotic transcripts, as well as the long noncoding RNA (lncRNA) MALAT1. ASOs were only effective within a narrow concentration range and were toxic at higher concentrations. Despite this drawback, quantitation of knockdown efficiency and the ability to degrade lncRNAs make ASOs a useful knockdown reagent in zebrafish.
Conflict of interest statement
Figures


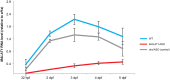
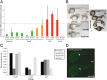
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

