Impact of Chromatin on HIV Replication
- PMID: 26437430
- PMCID: PMC4690024
- DOI: 10.3390/genes6040957
Impact of Chromatin on HIV Replication
Abstract
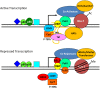
Chromatin influences Human Immunodeficiency Virus (HIV) integration and replication. This review highlights critical host factors that influence chromatin structure and organization and that also impact HIV integration, transcriptional regulation and latency. Furthermore, recent attempts to target chromatin associated factors to reduce the HIV proviral load are discussed.
Keywords: HIV; chromatin; integration; latency; transcription.
Figures
References
-
- Lewinski M.K., Bisgrove D., Shinn P., Chen H., Hoffmann C., Hannenhalli S., Verdin E., Berry C.C., Ecker J.R., Bushman F.D. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. - DOI - PMC - PubMed
-
- Shan L., Yang H.C., Rabi S.A., Bravo H.C., Shroff N.S., Irizarry R.A., Zhang H., Margolick J.B., Siliciano J.D., Siliciano R.F. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J. Virol. 2011;85:5384–5393. doi: 10.1128/JVI.02536-10. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

