Familial Dysautonomia (FD) Human Embryonic Stem Cell Derived PNS Neurons Reveal that Synaptic Vesicular and Neuronal Transport Genes Are Directly or Indirectly Affected by IKBKAP Downregulation
- PMID: 26437462
- PMCID: PMC4593545
- DOI: 10.1371/journal.pone.0138807
Familial Dysautonomia (FD) Human Embryonic Stem Cell Derived PNS Neurons Reveal that Synaptic Vesicular and Neuronal Transport Genes Are Directly or Indirectly Affected by IKBKAP Downregulation
Abstract
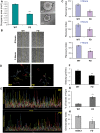
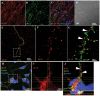
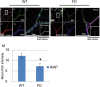
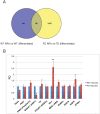
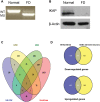
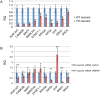
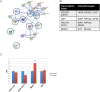
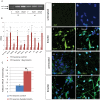
A splicing mutation in the IKBKAP gene causes Familial Dysautonomia (FD), affecting the IKAP protein expression levels and proper development and function of the peripheral nervous system (PNS). Here we found new molecular insights for the IKAP role and the impact of the FD mutation in the human PNS lineage by using a novel and unique human embryonic stem cell (hESC) line homozygous to the FD mutation originated by pre implantation genetic diagnosis (PGD) analysis. We found that IKBKAP downregulation during PNS differentiation affects normal migration in FD-hESC derived neural crest cells (NCC) while at later stages the PNS neurons show reduced intracellular colocalization between vesicular proteins and IKAP. Comparative wide transcriptome analysis of FD and WT hESC-derived neurons together with the analysis of human brains from FD and WT 12 weeks old embryos and experimental validation of the results confirmed that synaptic vesicular and neuronal transport genes are directly or indirectly affected by IKBKAP downregulation in FD neurons. Moreover we show that kinetin (a drug that corrects IKBKAP alternative splicing) promotes the recovery of IKAP expression and these IKAP functional associated genes identified in the study. Altogether, these results support the view that IKAP might be a vesicular like protein that might be involved in neuronal transport in hESC derived PNS neurons. This function seems to be mostly affected in FD-hESC derived PNS neurons probably reflecting some PNS neuronal dysfunction observed in FD.
Conflict of interest statement
Figures

Similar articles
-
Enriched population of PNS neurons derived from human embryonic stem cells as a platform for studying peripheral neuropathies.PLoS One. 2010 Feb 18;5(2):e9290. doi: 10.1371/journal.pone.0009290. PLoS One. 2010. PMID: 20174633 Free PMC article.
-
Involvement of IKAP in peripheral target innervation and in specific JNK and NGF signaling in developing PNS neurons.PLoS One. 2014 Nov 19;9(11):e113428. doi: 10.1371/journal.pone.0113428. eCollection 2014. PLoS One. 2014. PMID: 25409162 Free PMC article.
-
Proteasome inhibitors to alleviate aberrant IKBKAP mRNA splicing and low IKAP/hELP1 synthesis in familial dysautonomia.Neurobiol Dis. 2017 Jul;103:113-122. doi: 10.1016/j.nbd.2017.04.009. Epub 2017 Apr 9. Neurobiol Dis. 2017. PMID: 28404519
-
Animal and cellular models of familial dysautonomia.Clin Auton Res. 2017 Aug;27(4):235-243. doi: 10.1007/s10286-017-0438-2. Epub 2017 Jun 30. Clin Auton Res. 2017. PMID: 28667575 Free PMC article. Review.
-
Familial dysautonomia.Curr Opin Genet Dev. 2002 Jun;12(3):307-11. doi: 10.1016/s0959-437x(02)00303-9. Curr Opin Genet Dev. 2002. PMID: 12076674 Review.
Cited by
-
Dysregulated proteostasis network in neuronal diseases.Front Cell Dev Biol. 2023 Feb 24;11:1075215. doi: 10.3389/fcell.2023.1075215. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36910151 Free PMC article. Review.
-
Phosphatidylserine Ameliorates Neurodegenerative Symptoms and Enhances Axonal Transport in a Mouse Model of Familial Dysautonomia.PLoS Genet. 2016 Dec 20;12(12):e1006486. doi: 10.1371/journal.pgen.1006486. eCollection 2016 Dec. PLoS Genet. 2016. PMID: 27997532 Free PMC article.
-
BGP-15 prevents the death of neurons in a mouse model of familial dysautonomia.Proc Natl Acad Sci U S A. 2017 May 9;114(19):5035-5040. doi: 10.1073/pnas.1620212114. Epub 2017 Apr 24. Proc Natl Acad Sci U S A. 2017. PMID: 28439028 Free PMC article.
-
ATP-citrate lyase promotes axonal transport across species.Nat Commun. 2021 Oct 7;12(1):5878. doi: 10.1038/s41467-021-25786-y. Nat Commun. 2021. PMID: 34620845 Free PMC article.
-
Norepinephrine transporter defects lead to sympathetic hyperactivity in Familial Dysautonomia models.Nat Commun. 2022 Nov 17;13(1):7032. doi: 10.1038/s41467-022-34811-7. Nat Commun. 2022. PMID: 36396637 Free PMC article.
References
-
- Riley CM, Day RL, Greeley DM, Langford WS. Central autonomic dysfunction with defective lacrimation: report of five cases. Pediatrics. 1949;3:468–78. - PubMed
-
- Blumenfeld A, Slaugenhaupt SA, Axelrod FB, Lucente DE, Maayan C, Liebert CB, et al. Localization of the gene for familial dysautonomia on chromosome 9 and definition of DNA markers for genetic diagnosis. Nat Genet. 1993;4(2):160–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

