HES-Mediated Repression of Pten in Caenorhabditis elegans
- PMID: 26438299
- PMCID: PMC4683635
- DOI: 10.1534/g3.115.019463
HES-Mediated Repression of Pten in Caenorhabditis elegans
Abstract
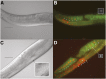
The hairy/enhancer-of-split (HES) group of transcription factors controls embryonic development, often by acting downstream of the Notch signaling pathway; however, little is known about postembryonic roles of these proteins. In Caenorhabditis elegans, the six proteins that make up the REF-1 family are considered to be HES orthologs that act in both Notch-dependent and Notch-independent pathways to regulate embryonic events. To further our understanding of how the REF-1 family works to coordinate postembryonic cellular events, we performed a functional characterization of the REF-1 family member, HLH-25. We show that, after embryogenesis, hlh-25 expression persists throughout every developmental stage, including dauer, into adulthood. Like animals that carry loss-of-function alleles in genes required for normal cell-cycle progression, the phenotypes of hlh-25 animals include reduced brood size, unfertilized oocytes, and abnormal gonad morphology. Using gene expression microarray, we show that the HLH-25 transcriptional network correlates with the phenotypes of hlh-25 animals and that the C. elegans Pten ortholog, daf-18, is one major hub in the network. Finally, we show that HLH-25 regulates C. elegans lifespan and dauer recovery, which correlates with a role in the transcriptional repression of daf-18 activity. Collectively, these data provide the first genetic evidence that HLH-25 may be a functional ortholog of mammalian HES1, which represses PTEN activity in mice and human cells.
Keywords: dauer recovery; gene expression microarray; gonad morphology; hairy/enhancer-of-split; unfertilized oocytes.
Copyright © 2015 Chou et al.
Figures

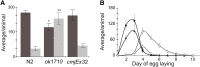


References
-
- Akazawa C., Ishibashi M., Shimizu C., Nakanishi S., Kageyama R., 1995. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem. 270: 8730–8738. - PubMed
-
- Alper S., Kenyon C., 2001. REF-1, a protein with two bHLH domains, alters the pattern of cell fusion in C. elegans by regulating Hox protein activity. Development 128: 1793–1804. - PubMed
-
- Axelson H., 2004. The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 204: 171–178. - PubMed
-
- Ben-Arie N., Hassan B. A., Bermingham N. A., Malicki D. M., Armstrong D., et al. , 2000. Functional conservation of atonal and Math1 in the CNS and PNS. Development 127: 1039–1048. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
