Genetics ignite focus on microglial inflammation in Alzheimer's disease
- PMID: 26438529
- PMCID: PMC4595327
- DOI: 10.1186/s13024-015-0048-1
Genetics ignite focus on microglial inflammation in Alzheimer's disease
Abstract
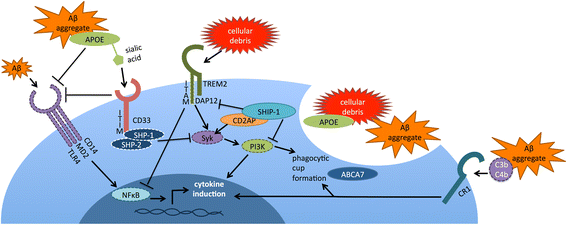
In the past five years, a series of large-scale genetic studies have revealed novel risk factors for Alzheimer's disease (AD). Analyses of these risk factors have focused attention upon the role of immune processes in AD, specifically microglial function. In this review, we discuss interpretation of genetic studies. We then focus upon six genes implicated by AD genetics that impact microglial function: TREM2, CD33, CR1, ABCA7, SHIP1, and APOE. We review the literature regarding the biological functions of these six proteins and their putative role in AD pathogenesis. We then present a model for how these factors may interact to modulate microglial function in AD.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

