Resistance to ketolide antibiotics by coordinated expression of rRNA methyltransferases in a bacterial producer of natural ketolides
- PMID: 26438831
- PMCID: PMC4620888
- DOI: 10.1073/pnas.1512090112
Resistance to ketolide antibiotics by coordinated expression of rRNA methyltransferases in a bacterial producer of natural ketolides
Abstract
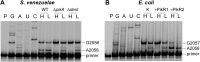
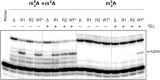
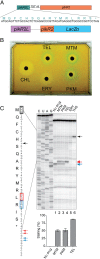
Ketolides are promising new antimicrobials effective against a broad range of Gram-positive pathogens, in part because of the low propensity of these drugs to trigger the expression of resistance genes. A natural ketolide pikromycin and a related compound methymycin are produced by Streptomyces venezuelae strain ATCC 15439. The producer avoids the inhibitory effects of its own antibiotics by expressing two paralogous rRNA methylase genes pikR1 and pikR2 with seemingly redundant functions. We show here that the PikR1 and PikR2 enzymes mono- and dimethylate, respectively, the N6 amino group in 23S rRNA nucleotide A2058. PikR1 monomethylase is constitutively expressed; it confers low resistance at low fitness cost and is required for ketolide-induced activation of pikR2 to attain high-level resistance. The regulatory mechanism controlling pikR2 expression has been evolutionary optimized for preferential activation by ketolide antibiotics. The resistance genes and the induction mechanism remain fully functional when transferred to heterologous bacterial hosts. The anticipated wide use of ketolide antibiotics could promote horizontal transfer of these highly efficient resistance genes to pathogens. Taken together, these findings emphasized the need for surveillance of pikR1/pikR2-based bacterial resistance and the preemptive development of drugs that can remain effective against the ketolide-specific resistance mechanism.
Keywords: antibiotics; ketolides; macrolides; resistance; ribosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

