Rapid Y degeneration and dosage compensation in plant sex chromosomes
- PMID: 26438872
- PMCID: PMC4620866
- DOI: 10.1073/pnas.1508454112
Rapid Y degeneration and dosage compensation in plant sex chromosomes
Abstract
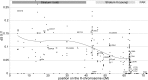
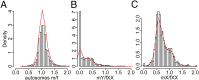
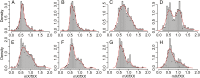
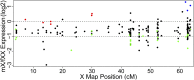
The nonrecombining regions of animal Y chromosomes are known to undergo genetic degeneration, but previous work has failed to reveal large-scale gene degeneration on plant Y chromosomes. Here, we uncover rapid and extensive degeneration of Y-linked genes in a plant species, Silene latifolia, that evolved sex chromosomes de novo in the last 10 million years. Previous transcriptome-based studies of this species missed unexpressed, degenerate Y-linked genes. To identify sex-linked genes, regardless of their expression, we sequenced male and female genomes of S. latifolia and integrated the genomic contigs with a high-density genetic map. This revealed that 45% of Y-linked genes are not expressed, and 23% are interrupted by premature stop codons. This contrasts with X-linked genes, in which only 1.3% of genes contained stop codons and 4.3% of genes were not expressed in males. Loss of functional Y-linked genes is partly compensated for by gene-specific up-regulation of X-linked genes. Our results demonstrate that the rate of genetic degeneration of Y-linked genes in S. latifolia is as fast as in animals, and that the evolutionary trajectories of sex chromosomes are similar in the two kingdoms.
Keywords: Y degeneration; dosage compensation; gene expression; plants; sex chromosome evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Renner SS, Ricklefs RE. Dioecy and its correlates in the flowering plants. Am J Bot. 1995;82(5):596–606.
-
- Charlesworth D. Plant sex chromosome evolution. J Exp Bot. 2013;64(2):405–420. - PubMed
-
- Westergaard M. The mechanism of sex determination in dioecious flowering plants. Adv Genet. 1958;9:217–281. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

