Cell Surface Proteomic Map of HIV Infection Reveals Antagonism of Amino Acid Metabolism by Vpu and Nef
- PMID: 26439863
- PMCID: PMC4608997
- DOI: 10.1016/j.chom.2015.09.003
Cell Surface Proteomic Map of HIV Infection Reveals Antagonism of Amino Acid Metabolism by Vpu and Nef
Abstract
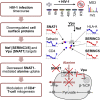
Critical cell surface immunoreceptors downregulated during HIV infection have previously been identified using non-systematic, candidate approaches. To gain a comprehensive, unbiased overview of how HIV infection remodels the T cell surface, we took a distinct, systems-level, quantitative proteomic approach. >100 plasma membrane proteins, many without characterized immune functions, were downregulated during HIV infection. Host factors targeted by the viral accessory proteins Vpu or Nef included the amino acid transporter SNAT1 and the serine carriers SERINC3/5. We focused on SNAT1, a β-TrCP-dependent Vpu substrate. SNAT1 antagonism was acquired by Vpu variants from the lineage of SIVcpz/HIV-1 viruses responsible for pandemic AIDS. We found marked SNAT1 induction in activated primary human CD4+ T cells, and used Consumption and Release (CoRe) metabolomics to identify alanine as an endogenous SNAT1 substrate required for T cell mitogenesis. Downregulation of SNAT1 therefore defines a unique paradigm of HIV interference with immunometabolism.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







Comment in
-
Attacking the Supply Lines: HIV-1 Restricts Alanine Uptake to Prevent T Cell Activation.Cell Host Microbe. 2015 Nov 11;18(5):514-7. doi: 10.1016/j.chom.2015.10.017. Cell Host Microbe. 2015. PMID: 26567503
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

