Quantitative Imaging of Gut Microbiota Spatial Organization
- PMID: 26439864
- PMCID: PMC4628835
- DOI: 10.1016/j.chom.2015.09.002
Quantitative Imaging of Gut Microbiota Spatial Organization
Abstract
Genomic technologies have significantly advanced our understanding of the composition and diversity of host-associated microbial populations. However, their spatial organization and functional interactions relative to the host have been more challenging to study. Here we present a pipeline for the assessment of intestinal microbiota localization within immunofluorescence images of fixed gut cross-sections that includes a flexible software package, BacSpace, for high-throughput quantification of microbial organization. Applying this pipeline to gnotobiotic and human microbiota-colonized mice, we demonstrate that elimination of microbiota-accessible carbohydrates (MACs) from the diet results in thinner mucus in the distal colon, increased proximity of microbes to the epithelium, and heightened expression of the inflammatory marker REG3β. Measurements of microbe-microbe proximity reveal that a MAC-deficient diet alters monophyletic spatial clustering. Furthermore, we quantify the invasion of Helicobacter pylori into the glands of the mouse stomach relative to host mitotic progenitor cells, illustrating the generalizability of this approach.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


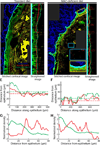


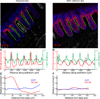
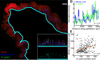
Comment in
-
Bacteria Come into Focus: New Tools for Visualizing the Microbiota.Cell Host Microbe. 2015 Oct 14;18(4):392-4. doi: 10.1016/j.chom.2015.10.004. Cell Host Microbe. 2015. PMID: 26468742
References
-
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10771–10776. - PMC - PubMed
-
- Bayley GVHJM. The “Effective” Number of Independent Observations in an Autocorrelated Time Series. Supplement to the Journal of the Royal Statistical Society. 1946;8:184–197.
-
- Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, van der Sluis M, Lu P, Van Goudoever JB, Wells JM, Dekker J, et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3beta, Reg3gamma and angiogenin-4. PloS one. 2012;7:e38798. - PMC - PubMed
-
- Burkitt DP. Epidemiology of large bowel disease: the role of fibre. The Proceedings of the Nutrition Society. 1973;32:145–149. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

