Divergent Evolution of the Transcriptional Network Controlled by Snf1-Interacting Protein Sip4 in Budding Yeasts
- PMID: 26440109
- PMCID: PMC4634231
- DOI: 10.1371/journal.pone.0139464
Divergent Evolution of the Transcriptional Network Controlled by Snf1-Interacting Protein Sip4 in Budding Yeasts
Abstract
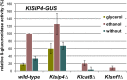
Cellular responses to starvation are of ancient origin since nutrient limitation has always been a common challenge to the stability of living systems. Hence, signaling molecules involved in sensing or transducing information about limiting metabolites are highly conserved, whereas transcription factors and the genes they regulate have diverged. In eukaryotes the AMP-activated protein kinase (AMPK) functions as a central regulator of cellular energy homeostasis. The yeast AMPK ortholog SNF1 controls the transcriptional network that counteracts carbon starvation conditions by regulating a set of transcription factors. Among those Cat8 and Sip4 have overlapping DNA-binding specificity for so-called carbon source responsive elements and induce target genes upon SNF1 activation. To analyze the evolution of the Cat8-Sip4 controlled transcriptional network we have compared the response to carbon limitation of Saccharomyces cerevisiae to that of Kluyveromyces lactis. In high glucose, S. cerevisiae displays tumor cell-like aerobic fermentation and repression of respiration (Crabtree-positive) while K. lactis has a respiratory-fermentative life-style, respiration being regulated by oxygen availability (Crabtree-negative), which is typical for many yeasts and for differentiated higher cells. We demonstrate divergent evolution of the Cat8-Sip4 network and present evidence that a role of Sip4 in controlling anabolic metabolism has been lost in the Saccharomyces lineage. We find that in K. lactis, but not in S. cerevisiae, the Sip4 protein plays an essential role in C2 carbon assimilation including induction of the glyoxylate cycle and the carnitine shuttle genes. Induction of KlSIP4 gene expression by KlCat8 is essential under these growth conditions and a primary function of KlCat8. Both KlCat8 and KlSip4 are involved in the regulation of lactose metabolism in K. lactis. In chromatin-immunoprecipitation experiments we demonstrate binding of both, KlSip4 and KlCat8, to selected CSREs and provide evidence that KlSip4 counteracts KlCat8-mediated transcription activation by competing for binding to some but not all CSREs. The finding that the hierarchical relationship of these transcription factors differs between K. lactis and S. cerevisiae and that the sets of target genes have diverged contributes to explaining the phenotypic differences in metabolic life-style.
Conflict of interest statement
Figures






References
-
- Towler MC, Hardie DG (2007) AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res 100: 328–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

