The human gut virome: a multifaceted majority
- PMID: 26441861
- PMCID: PMC4566309
- DOI: 10.3389/fmicb.2015.00918
The human gut virome: a multifaceted majority
Abstract
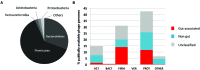
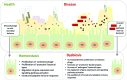
Here, we outline our current understanding of the human gut virome, in particular the phage component of this ecosystem, highlighting progress, and challenges in viral discovery in this arena. We reveal how developments in high-throughput sequencing technologies and associated data analysis methodologies are helping to illuminate this abundant 'biological dark matter.' Current evidence suggests that the human gut virome is a highly individual but temporally stable collective, dominated by phages exhibiting a temperate lifestyle. This viral community also appears to encode a surprisingly rich functional repertoire that confers a range of attributes to their bacterial hosts, ranging from bacterial virulence and pathogenesis to maintaining host-microbiome stability and community resilience. Despite the significant advances in our understanding of the gut virome in recent years, it is clear that we remain in a period of discovery and revelation, as new methods and technologies begin to provide deeper understanding of the inherent ecological characteristics of this viral ecosystem. As our understanding increases, the nature of the multi-partite interactions occurring between host and microbiome will become clearer, helping us to more rationally define the concepts and principles that will underpin approaches to using human gut virome components for medical or biotechnological applications.
Keywords: bacteriophage; dysbiosis; human gut microbiome; metagenomics; phage-encoded functions; virus-like particles.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

