Nuclear localization of the dehydrin OpsDHN1 is determined by histidine-rich motif
- PMID: 26442018
- PMCID: PMC4561349
- DOI: 10.3389/fpls.2015.00702
Nuclear localization of the dehydrin OpsDHN1 is determined by histidine-rich motif
Abstract
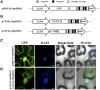
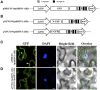
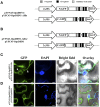
The cactus OpsDHN1 dehydrin belongs to a large family of disordered and highly hydrophilic proteins known as Late Embryogenesis Abundant (LEA) proteins, which accumulate during the late stages of embryogenesis and in response to abiotic stresses. Herein, we present the in vivo OpsDHN1 subcellular localization by N-terminal GFP translational fusion; our results revealed a cytoplasmic and nuclear localization of the GFP::OpsDHN1 protein in Nicotiana benthamiana epidermal cells. In addition, dimer assembly of OpsDHN1 in planta using a Bimolecular Fluorescence Complementation (BiFC) approach was demonstrated. In order to understand the in vivo role of the histidine-rich motif, the OpsDHN1-ΔHis version was produced and assayed for its subcellular localization and dimer capability by GFP fusion and BiFC assays, respectively. We found that deletion of the OpsDHN1 histidine-rich motif restricted its localization to cytoplasm, but did not affect dimer formation. In addition, the deletion of the S-segment in the OpsDHN1 protein affected its nuclear localization. Our data suggest that the deletion of histidine-rich motif and S-segment show similar effects, preventing OpsDHN1 from getting into the nucleus. Based on these results, the histidine-rich motif is proposed as a targeting element for OpsDHN1 nuclear localization.
Keywords: BiFC; dehydrin; histidine-rich motif; homodimer; nuclear/cytoplasmic localization.
Figures


References
-
- Close T. J. (1996). Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 97, 795–803. 10.1111/j.1399-3054.1996.tb00546.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

