Genetic variation of temperature-regulated curd induction in cauliflower: elucidation of floral transition by genome-wide association mapping and gene expression analysis
- PMID: 26442034
- PMCID: PMC4564693
- DOI: 10.3389/fpls.2015.00720
Genetic variation of temperature-regulated curd induction in cauliflower: elucidation of floral transition by genome-wide association mapping and gene expression analysis
Abstract
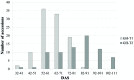
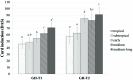
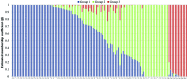
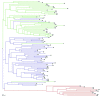
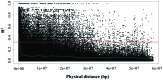
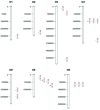
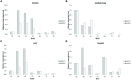
Cauliflower (Brassica oleracea var. botrytis) is a vernalization-responsive crop. High ambient temperatures delay harvest time. The elucidation of the genetic regulation of floral transition is highly interesting for a precise harvest scheduling and to ensure stable market supply. This study aims at genetic dissection of temperature-dependent curd induction in cauliflower by genome-wide association studies and gene expression analysis. To assess temperature-dependent curd induction, two greenhouse trials under distinct temperature regimes were conducted on a diversity panel consisting of 111 cauliflower commercial parent lines, genotyped with 14,385 SNPs. Broad phenotypic variation and high heritability (0.93) were observed for temperature-related curd induction within the cauliflower population. GWA mapping identified a total of 18 QTL localized on chromosomes O1, O2, O3, O4, O6, O8, and O9 for curding time under two distinct temperature regimes. Among those, several QTL are localized within regions of promising candidate flowering genes. Inferring population structure and genetic relatedness among the diversity set assigned three main genetic clusters. Linkage disequilibrium (LD) patterns estimated global LD extent of r(2) = 0.06 and a maximum physical distance of 400 kb for genetic linkage. Transcriptional profiling of flowering genes FLOWERING LOCUS C (BoFLC) and VERNALIZATION 2 (BoVRN2) was performed, showing increased expression levels of BoVRN2 in genotypes with faster curding. However, functional relevance of BoVRN2 and BoFLC2 could not consistently be supported, which probably suggests to act facultative and/or might evidence for BoVRN2/BoFLC-independent mechanisms in temperature-regulated floral transition in cauliflower. Genetic insights in temperature-regulated curd induction can underpin genetically informed phenology models and benefit molecular breeding strategies toward the development of thermo-tolerant cultivars.
Keywords: cauliflower; curd induction; genome-wide association study (GWAS); linkage disequilibrium (LD); quantitative trait loci (QTL); single nucleotide polymorphism (SNP); transcriptional profiling; vernalization.
Figures

References
-
- Abrol D. P. (2012). Pollination Biology: Biodiversity Conservation and Agricultural Production. New York, NY: Springer.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

