Genome-wide gene expression perturbation induced by loss of C2 chromosome in allotetraploid Brassica napus L
- PMID: 26442076
- PMCID: PMC4585227
- DOI: 10.3389/fpls.2015.00763
Genome-wide gene expression perturbation induced by loss of C2 chromosome in allotetraploid Brassica napus L
Abstract
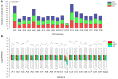
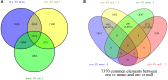
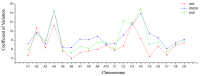
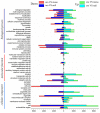
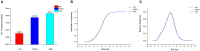
Aneuploidy with loss of entire chromosomes from normal complement disrupts the balanced genome and is tolerable only by polyploidy plants. In this study, the monosomic and nullisomic plants losing one or two copies of C2 chromosome from allotetraploid Brassica napus L. (2n = 38, AACC) were produced and compared for their phenotype and transcriptome. The monosomics gave a plant phenotype very similar to the original donor, but the nullisomics had much smaller stature and also shorter growth period. By the comparative analyses on the global transcript profiles with the euploid donor, genome-wide alterations in gene expression were revealed in two aneuploids, and their majority of differentially expressed genes (DEGs) resulted from the trans-acting effects of the zero and one copy of C2 chromosome. The higher number of up-regulated genes than down-regulated genes on other chromosomes suggested that the genome responded to the C2 loss via enhancing the expression of certain genes. Particularly, more DEGs were detected in the monosomics than nullisomics, contrasting with their phenotypes. The gene expression of the other chromosomes was differently affected, and several dysregulated domains in which up- or downregulated genes obviously clustered were identifiable. But the mean gene expression (MGE) for homoeologous chromosome A2 reduced with the C2 loss. Some genes and their expressions on C2 were correlated with the phenotype deviations in the aneuploids. These results provided new insights into the transcriptomic perturbation of the allopolyploid genome elicited by the loss of individual chromosome.
Keywords: Brassica napus; aneuploids; monosomy; nullisomy; transcriptome.
Figures

References
-
- Blakeslee A. F. (1922). Variations in Datura due to changes in chromosome number. Am. Nat. 56, 16–31. 10.1086/279845 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

