A survey about methods dedicated to epistasis detection
- PMID: 26442103
- PMCID: PMC4564769
- DOI: 10.3389/fgene.2015.00285
A survey about methods dedicated to epistasis detection
Abstract
During the past decade, findings of genome-wide association studies (GWAS) improved our knowledge and understanding of disease genetics. To date, thousands of SNPs have been associated with diseases and other complex traits. Statistical analysis typically looks for association between a phenotype and a SNP taken individually via single-locus tests. However, geneticists admit this is an oversimplified approach to tackle the complexity of underlying biological mechanisms. Interaction between SNPs, namely epistasis, must be considered. Unfortunately, epistasis detection gives rise to analytic challenges since analyzing every SNP combination is at present impractical at a genome-wide scale. In this review, we will present the main strategies recently proposed to detect epistatic interactions, along with their operating principle. Some of these methods are exhaustive, such as multifactor dimensionality reduction, likelihood ratio-based tests or receiver operating characteristic curve analysis; some are non-exhaustive, such as machine learning techniques (random forests, Bayesian networks) or combinatorial optimization approaches (ant colony optimization, computational evolution system).
Keywords: biological data mining; complex disease; epistasis detection; feature selection; genome-wide association study.
Figures
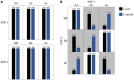
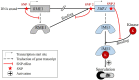
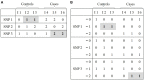
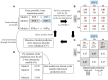


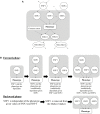
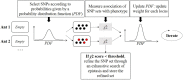
References
-
- Agresti A. (2002). Categorical Data Analysis, 2nd Edn. Hoboken, NJ: John Wiley & Sons, Inc.
-
- Aliferis C. F., Statnikov A., Tsamardinos I., Mani S., Koutsoukos X. D. (2010a). Local causal and markov blanket induction for causal discovery and feature selection for classification part I: algorithms and empirical evaluation. J. Mach. Learn. Res. 11, 171–234.
-
- Aliferis C. F., Statnikov A., Tsamardinos I., Mani S., Koutsoukos X. D. (2010b). Local Causal and markov blanket induction for causal discovery and feature selection for classification part II: analysis and extensions. J. Mach. Learn. Res. 11, 235-284.
-
- Bateson W. (1909). Mendel's Principles of Heredity. Cambridge, UK: Cambridge University Press.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

