Complete genome sequence of Geobacillus thermoglucosidasius C56-YS93, a novel biomass degrader isolated from obsidian hot spring in Yellowstone National Park
- PMID: 26442136
- PMCID: PMC4593210
- DOI: 10.1186/s40793-015-0031-z
Complete genome sequence of Geobacillus thermoglucosidasius C56-YS93, a novel biomass degrader isolated from obsidian hot spring in Yellowstone National Park
Abstract
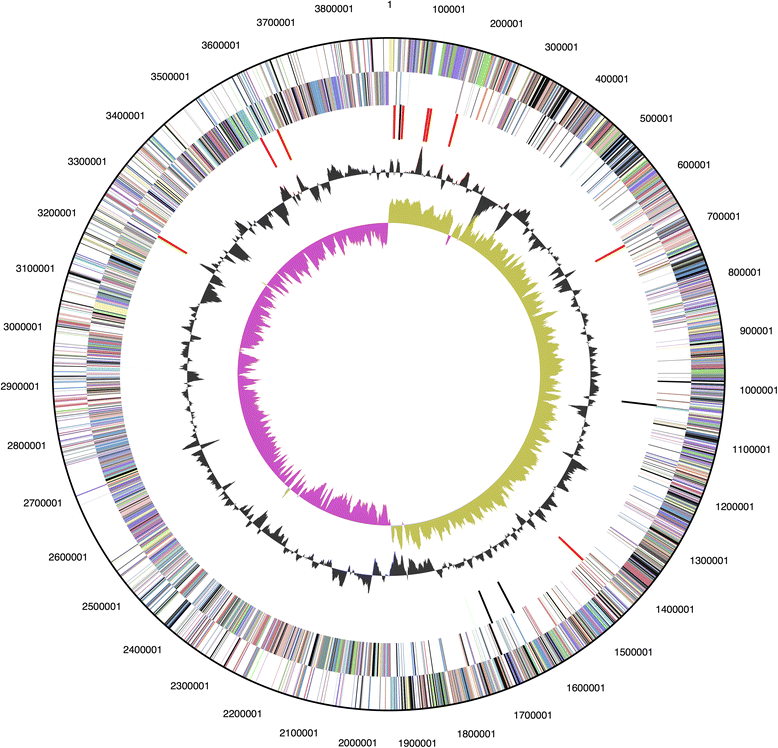
Geobacillus thermoglucosidasius C56-YS93 was one of several thermophilic organisms isolated from Obsidian Hot Spring, Yellowstone National Park, Montana, USA under permit from the National Park Service. Comparison of 16 S rRNA sequences confirmed the classification of the strain as a G. thermoglucosidasius species. The genome was sequenced, assembled, and annotated by the DOE Joint Genome Institute and deposited at the NCBI in December 2011 (CP002835). The genome of G. thermoglucosidasius C56-YS93 consists of one circular chromosome of 3,893,306 bp and two circular plasmids of 80,849 and 19,638 bp and an average G + C content of 43.93 %. G. thermoglucosidasius C56-YS93 possesses a xylan degradation cluster not found in the other G. thermoglucosidasius sequenced strains. This cluster appears to be related to the xylan degradation cluster found in G. stearothermophilus. G. thermoglucosidasius C56-YS93 possesses two plasmids not found in the other two strains. One plasmid contains a novel gene cluster coding for proteins involved in proline degradation and metabolism, the other contains a collection of mostly hypothetical proteins.
Keywords: Biomass; Geobacillus thermoglucosidasius C56-YS93; Hot springs; Prophage; Xylan.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

