Analytics for Metabolic Engineering
- PMID: 26442249
- PMCID: PMC4561385
- DOI: 10.3389/fbioe.2015.00135
Analytics for Metabolic Engineering
Abstract
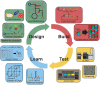
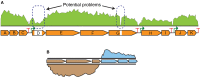
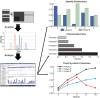
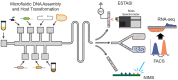
Realizing the promise of metabolic engineering has been slowed by challenges related to moving beyond proof-of-concept examples to robust and economically viable systems. Key to advancing metabolic engineering beyond trial-and-error research is access to parts with well-defined performance metrics that can be readily applied in vastly different contexts with predictable effects. As the field now stands, research depends greatly on analytical tools that assay target molecules, transcripts, proteins, and metabolites across different hosts and pathways. Screening technologies yield specific information for many thousands of strain variants, while deep omics analysis provides a systems-level view of the cell factory. Efforts focused on a combination of these analyses yield quantitative information of dynamic processes between parts and the host chassis that drive the next engineering steps. Overall, the data generated from these types of assays aid better decision-making at the design and strain construction stages to speed progress in metabolic engineering research.
Keywords: RNA-seq; high-throughput screening; metabolic engineering; metabolomics; microfluidics; proteomics.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

