The Microbiome in Populations with a Low and High Prevalence of Caries
- PMID: 26442950
- PMCID: PMC4700664
- DOI: 10.1177/0022034515609554
The Microbiome in Populations with a Low and High Prevalence of Caries
Abstract
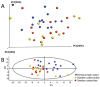
The oral microbiota was compared between Romanian adolescents with a high prevalence of caries and no dental care and Swedish caries-active and caries-free adolescents in caries prevention programs and with a low prevalence of caries. Biofilm samples were analyzed by FLX+ pyrosequencing of the V1 to V4 hypervariable regions of the 16S rRNA gene and polymerase chain reaction (PCR)/quantitative PCR (qPCR) for Streptococcus mutans and Streptococcus sobrinus. Sequences obtained blasted to 9 phyla, 66 genera, and 401 human oral taxa (HOT) in the 16S rRNA Human Oral Microbiome Database, of which 295 were represented by ≥20 sequences. The Romanian adolescents had more sequences in Firmicutes and fewer in Actinobacteria phyla and more sequences in the genera Bacteroidetes [G-3], Porphyromonas, Abiotrophia, Filifactor, Peptostreptococcaceae [11][G-4], Pseudoramibacter, Streptococcus, and Neisseria and fewer in Actinomyces, Selenomonas, Veillonella, Campylobacter, and TM7 [G-1] than the Swedish groups. Multivariate modeling employing HOT, S. sobrinus and S. mutans (PCR/qPCR), and sugar snacks separated Romanian from Swedish adolescents. The Romanian adolescents' microbiota was characterized by a panel of streptococci, including S. mutans, S. sobrinus, and Streptococcus australis, and Alloprevotella, Leptotrichia, Neisseria, Porphyromonas, and Prevotella. The Swedish adolescents were characterized by sweet snacks, and those with caries activity were also characterized by Prevotella, Actinomyces, and Capnocytophaga species and those free of caries by Actinomyces, Prevotella, Selenomonas, Streptococcus, and Mycoplasma. Eight species including Streptococcus mitis and Streptococcus species HOT070 were prevalent in Romanian and Swedish caries-active subjects but not caries-free subjects. In conclusion, S. mutans and S. sobrinus correlated with Romanian adolescents with caries and with limited access to dental care, whereas S. mutans and S. sobrinus were detected infrequently in Swedish adolescents in dental care programs. Swedish caries-active adolescents were typically colonized by Actinomyces, Selenomonas, Prevotella, and Capnocytophaga. Hence, the role of mutans streptococci as a primary caries pathogen appears less pronounced in populations with prevention programs compared to populations lacking caries treatment and prevention strategies.
Keywords: Romania; Sweden; adolescents; mutans streptococci; oral microbiota; pyrosequencing.
© International & American Associations for Dental Research 2015.
Conflict of interest statement
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures


References
-
- Do LG. 2012. Distribution of caries in children variations between and within populations. J Dent Res. 91(6):536–543. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

