Arabidopsis ABA-Activated Kinase MAPKKK18 is Regulated by Protein Phosphatase 2C ABI1 and the Ubiquitin-Proteasome Pathway
- PMID: 26443375
- PMCID: PMC4675898
- DOI: 10.1093/pcp/pcv146
Arabidopsis ABA-Activated Kinase MAPKKK18 is Regulated by Protein Phosphatase 2C ABI1 and the Ubiquitin-Proteasome Pathway
Abstract
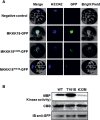
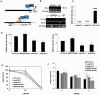
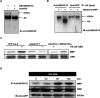
Phosphorylation and dephosphorylation events play an important role in the transmission of the ABA signal. Although SnRK2 [sucrose non-fermenting1-related kinase2] protein kinases and group A protein phosphatase type 2C (PP2C)-type phosphatases constitute the core ABA pathway, mitogen-activated protein kinase (MAPK) pathways are also involved in plant response to ABA. However, little is known about the interplay between MAPKs and PP2Cs or SnRK2 in the regulation of ABA pathways. In this study, an effort was made to elucidate the role of MAP kinase kinase kinase18 (MKKK18) in relation to ABA signaling and response. The MKKK18 knockout lines showed more vigorous root growth, decreased abaxial stomatal index and increased stomatal aperture under normal growth conditions, compared with the control wild-type Columbia line. In addition to transcriptional regulation of the MKKK18 promoter by ABA, we demonstrated using in vitro and in vivo kinase assays that the kinase activity of MKKK18 was regulated by ABA. Analysis of the cellular localization of MKKK18 showed that the active kinase was targeted specifically to the nucleus. Notably, we identified abscisic acid insensitive 1 (ABI1) PP2C as a MKKK18-interacting protein, and demonstrated that ABI1 inhibited its activity. Using a cell-free degradation assay, we also established that MKKK18 was unstable and was degraded by the proteasome pathway. The rate of MKKK18 degradation was delayed in the ABI1 knockout line. Overall, we provide evidence that ABI1 regulates the activity and promotes proteasomal degradation of MKKK18.
Keywords: ABA signaling; ABI1 PP2C; Arabidopsis thaliana; MAP kinase cascade; MKKK18; Proteasome.
© The Author 2015. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Figures





References
-
- Bate N., Twell D. (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 37: 859–869. - PubMed
-
- Cai G., Wang G., Wang L., Pan J., Liu Y., Li D. (2014) ZmMKK1, a novel group A mitogen-activated protein kinase kinase gene in maize, conferred chilling stress tolerance and was involved in pathogen defense in transgenic tobacco. Plant Sci. 214: 57–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

