Peste des petits ruminants
- PMID: 26443889
- PMCID: PMC4655833
- DOI: 10.1016/j.vetmic.2015.08.009
Peste des petits ruminants
Abstract
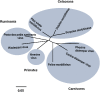
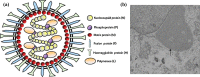
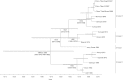
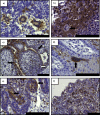
Peste des petits ruminants virus causes a highly infectious disease of small ruminants that is endemic across Africa, the Middle East and large regions of Asia. The virus is considered to be a major obstacle to the development of sustainable agriculture across the developing world and has recently been targeted by the World Organisation for Animal Health (OIE) and the Food and Agriculture Organisation (FAO) for eradication with the aim of global elimination of the disease by 2030. Fundamentally, the vaccines required to successfully achieve this goal are currently available, but the availability of novel vaccine preparations to also fulfill the requisite for differentiation between infected and vaccinated animals (DIVA) may reduce the time taken and the financial costs of serological surveillance in the later stages of any eradication campaign. Here, we overview what is currently known about the virus, with reference to its origin, updated global circulation, molecular evolution, diagnostic tools and vaccines currently available to combat the disease. Further, we comment on recent developments in our knowledge of various recombinant vaccines and on the potential for the development of novel multivalent vaccines for small ruminants.
Keywords: Control and eradication; Country-wise virus circulation; Live attenuated vaccine; Molecular evolution; PPR; Pathogenesis; Potential DIVA and multivalent vaccine; Reverse genetics.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Abd El-Rahim I.H., Sharawi S.S., Barakat M.R., El-Nahas E.M. An outbreak of peste des petits ruminants in migratory flocks of sheep and goats in Egypt in 2006. Rev. Sci. Technol. 2010;29:655–662. - PubMed
-
- Abu Elzein E.M.E., Housawi F.M.T., Bashareek Y., Gameel A.A., Al-Afaleq A.I., Anderson E. Severe PPR infection in gazelles kept under semi-free range conditions. J. Vet. Med. B. 2004;51:68–71. - PubMed
-
- Abubakar M., Arshed M.J., Hussain M., Ali Q. Evidence of peste des petits ruminants in serology of sheep and goats from Sindh, Pakistan. Transbound. Emerg. Dis. 2011;58:152–156. - PubMed
-
- Adombi C.M., Lelenta M., Lamien C.E., Shamaki D., Koffi Y.M., Traore A., Silber R., Couacy-Hymann E., Bodjo S.C., Djaman J.A., Luckins A.G., Diallo A. Monkey CV1 cell line expressing the sheep–goat SLAM protein: a highly sensitive cell line for the isolation of peste des petits ruminants virus from pathological specimens. J. Virol. Methods. 2011;173:306–313. - PMC - PubMed
-
- Adu F.D., Joannis T., Nwosuh E., Abegunde A. Pathogenicity of attenuated peste des petits ruminants virus in sheep and goats. Rev. Elev. Med. Vet. Pays Trop. 1990;43:23–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/J020478/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I026138/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L004801/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L013657/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- HH009485/1/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

