Evolutionary divergence of core and post-translational circadian clock genes in the pitcher-plant mosquito, Wyeomyia smithii
- PMID: 26444857
- PMCID: PMC4594641
- DOI: 10.1186/s12864-015-1937-y
Evolutionary divergence of core and post-translational circadian clock genes in the pitcher-plant mosquito, Wyeomyia smithii
Abstract
Background: Internal circadian (circa, about; dies, day) clocks enable organisms to maintain adaptive timing of their daily behavioral activities and physiological functions. Eukaryotic clocks consist of core transcription-translation feedback loops that generate a cycle and post-translational modifiers that maintain that cycle at about 24 h. We use the pitcher-plant mosquito, Wyeomyia smithii (subfamily Culicini, tribe Sabethini), to test whether evolutionary divergence of the circadian clock genes in this species, relative to other insects, has involved primarily genes in the core feedback loops or the post-translational modifiers. Heretofore, there is no reference transcriptome or genome sequence for any mosquito in the tribe Sabethini, which includes over 375 mainly circumtropical species.
Methods: We sequenced, assembled and annotated the transcriptome of W. smithii containing nearly 95 % of conserved single-copy orthologs in animal genomes. We used the translated contigs and singletons to determine the average rates of circadian clock-gene divergence in W. smithii relative to three other mosquito genera, to Drosophila, to the butterfly, Danaus, and to the wasp, Nasonia.
Results: Over 1.08 million cDNA sequence reads were obtained consisting of 432.5 million nucleotides. Their assembly produced 25,904 contigs and 54,418 singletons of which 62 % and 28 % are annotated as protein-coding genes, respectively, sharing homology with other animal proteomes.
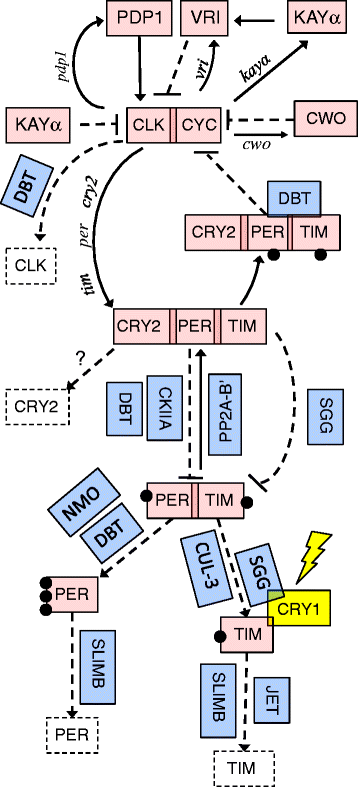
Discussion: The W. smithii transcriptome includes all nine circadian transcription-translation feedback-loop genes and all eight post-translational modifier genes we sought to identify (Fig. 1). After aligning translated W. smithii contigs and singletons from this transcriptome with other insects, we determined that there was no significant difference in the average divergence of W. smithii from the six other taxa between the core feedback-loop genes and post-translational modifiers.
Conclusions: The characterized transcriptome is sufficiently complete and of sufficient quality to have uncovered all of the insect circadian clock genes we sought to identify (Fig. 1). Relative divergence does not differ between core feedback-loop genes and post-translational modifiers of those genes in a Sabethine species (W. smithii) that has experienced a continual northward dispersal into temperate regions of progressively longer summer day lengths as compared with six other insect taxa. An associated microarray platform derived from this work will enable the investigation of functional genomics of circadian rhythmicity, photoperiodic time measurement, and diapause along a photic and seasonal geographic gradient.
Figures




Similar articles
-
Clock-talk: have we forgotten about geographic variation?J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2024 Jul;210(4):649-666. doi: 10.1007/s00359-023-01643-9. Epub 2023 Jun 16. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2024. PMID: 37322375 Free PMC article.
-
Genetic correlations and the evolution of photoperiodic time measurement within a local population of the pitcher-plant mosquito, Wyeomyia smithii.Heredity (Edinb). 2012 May;108(5):473-9. doi: 10.1038/hdy.2011.108. Epub 2011 Nov 9. Heredity (Edinb). 2012. PMID: 22072069 Free PMC article.
-
Evolutionary divergence of circadian and photoperiodic phenotypes in the pitcher-plant mosquito, Wyeomyia smithii.J Biol Rhythms. 2006 Apr;21(2):132-9. doi: 10.1177/0748730406286320. J Biol Rhythms. 2006. PMID: 16603677
-
Natural Variation and Genetics of Photoperiodism in Wyeomyia smithii.Adv Genet. 2017;99:39-71. doi: 10.1016/bs.adgen.2017.09.002. Epub 2017 Oct 12. Adv Genet. 2017. PMID: 29050554 Review.
-
Evolutionary links between circadian clocks and photoperiodic diapause in insects.Integr Comp Biol. 2013 Jul;53(1):131-43. doi: 10.1093/icb/ict023. Epub 2013 Apr 24. Integr Comp Biol. 2013. PMID: 23615363 Free PMC article. Review.
Cited by
-
Comparative Transcriptomics Reveals Key Gene Expression Differences between Diapausing and Non-Diapausing Adults of Culex pipiens.PLoS One. 2016 Apr 29;11(4):e0154892. doi: 10.1371/journal.pone.0154892. eCollection 2016. PLoS One. 2016. PMID: 27128578 Free PMC article.
-
Keeping time without a spine: what can the insect clock teach us about seasonal adaptation?Philos Trans R Soc Lond B Biol Sci. 2017 Nov 19;372(1734):20160257. doi: 10.1098/rstb.2016.0257. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28993500 Free PMC article. Review.
-
Evolutionary transition from blood feeding to obligate nonbiting in a mosquito.Proc Natl Acad Sci U S A. 2018 Jan 30;115(5):1009-1014. doi: 10.1073/pnas.1717502115. Epub 2017 Dec 18. Proc Natl Acad Sci U S A. 2018. PMID: 29255013 Free PMC article.
-
Evidence that microRNAs are part of the molecular toolkit regulating adult reproductive diapause in the mosquito, Culex pipiens.PLoS One. 2018 Nov 29;13(11):e0203015. doi: 10.1371/journal.pone.0203015. eCollection 2018. PLoS One. 2018. PMID: 30496183 Free PMC article.
-
Daily Rhythms in Mosquitoes and Their Consequences for Malaria Transmission.Insects. 2016 Apr 14;7(2):14. doi: 10.3390/insects7020014. Insects. 2016. PMID: 27089370 Free PMC article. Review.
References
-
- Hopkins AD. Bioclimatics: a science of life and climate relations. USDA Miscellaneous Publication No. 280. Washington (DC): United States Government Printing Office; 1938.
-
- MacArthur RH. Geographical ecology. Princeton (NJ): Princeton University Press; 1984.
-
- Hoffmann AA, Parsons PA. Extreme environmental change and evolution. Cambridge (UK): Cambridge University Press; 1997.
-
- Angilletta MJ., Jr . Thermal adaptation: a theoretical and empirical synthesis. Oxford (UK): Oxford University Press; 2009.
-
- Gilbert SF, Epel D. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland (MA): Sinauer Associates; 2009.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

