RBM10 Modulates Apoptosis and Influences TNF-α Gene Expression
- PMID: 26446321
- PMCID: PMC4583097
- DOI: 10.4137/JCD.S9073
RBM10 Modulates Apoptosis and Influences TNF-α Gene Expression
Abstract
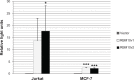
Recent evidence suggests that protein encoded by the RNA Binding Motif 10 (RBM10) gene has the ability to modulate apoptosis. The objective of this study was to test this hypothesis by manipulating RBM10 expression levels and examining the downstream consequences. The results showed that transient overexpression of RBM10 correlated with significantly elevated levels of tumour necrosis factor alpha (TNF-α) mRNA and soluble TNF-α (sTNF-α) protein, and increased apoptosis (phosphatidyl serine exposure on the outer cell membrane and nuclear condensation). Stable RNA interference-mediated RBM10 knockdown clones were less susceptible to TNF-α-mediated apoptosis, and had decreased sTNF-α protein levels. Elevated levels of TNF-α associated with RBM10 overexpression resulted from increased TNF-α transcription, not TNF-α mRNA stabilization. These results suggest that RBM10 has the ability to modulate apoptosis, and that it does so via a mechanism involving alterations to TNFR super family-mediated signaling. These data provide the first direct evidence that human RBM10 can function as an apoptosis modulator and cytokine expression regulator.
Keywords: RBM10; RBM5; TNF-alpha; apoptosis; cancer; transcription.
Figures




References
-
- Timmer T, Terpstra P, van den Berg A, et al. An evolutionary rearrangement of the Xp11.3–11.23 region in 3p21.3, a region frequently deleted in a variety of cancers. Genomics. 1999;60:238–240. - PubMed
-
- Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell. 2008;32:81–95. - PubMed
-
- Martinez-Arribas F, Agudo D, Pollan M, et al. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem. 2006;97:1275–1282. - PubMed
-
- Martin-Garabato E, Martinez-Arribas F, Pollan M, Lucas AR, Sanchez J, Schneider J. The small variant of the apoptosis-associated X-chromosome RBM10 gene is co-expressed with caspase-3 in breast cancer. Cancer Genomics Proteomics. 2008;5:169–73. - PubMed
LinkOut - more resources
Full Text Sources

