Characterizing the Murine Leukemia Virus Envelope Glycoprotein Membrane-Spanning Domain for Its Roles in Interface Alignment and Fusogenicity
- PMID: 26446598
- PMCID: PMC4665228
- DOI: 10.1128/JVI.01901-15
Characterizing the Murine Leukemia Virus Envelope Glycoprotein Membrane-Spanning Domain for Its Roles in Interface Alignment and Fusogenicity
Abstract
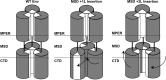
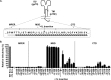
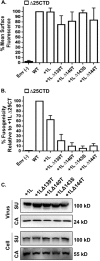
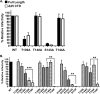
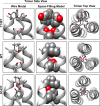
The membrane-proximal region of murine leukemia virus envelope (Env) is a critical modulator of its functionality. We have previously shown that the insertion of one amino acid (+1 leucine) within the membrane-spanning domain (MSD) abolished protein functionality in infectivity assays. However, functionality could be restored to this +1 leucine mutant by either inserting two additional amino acids (+3 leucine) or by deleting the cytoplasmic tail domain (CTD) in the +1 leucine background. We inferred that the ectodomain and CTD have protein interfaces that have to be in alignment for Env to be functional. Here, we made single residue deletions to the Env mutant with the +1 leucine insertion to restore the interface alignment (gain of functionality) and therefore define the boundaries of the two interfaces. We identified the glycine-proline pairs near the N terminus (positions 147 and 148) and the C terminus (positions 159 and 160) of the MSD as being the boundaries of the two interfaces. Deletions between these pairs restored function, but deletions outside of them did not. In addition, the vast majority of the single residue deletions regained function if the CTD was deleted. The exceptions were four hydroxyl-containing amino acid residues (T139, T140, S143, and T144) that reside in the ectodomain interface and the proline at position 148, which were all indispensable for functionality. We hypothesize that the hydroxyl-containing residues at positions T139 and S143 could be a driving force for stabilizing the ectodomain interface through formation of a hydrogen-bonding network.
Importance: The membrane-proximal external region (MPER) and membrane-spanning domains (MSDs) of viral glycoproteins have been shown to be critical for regulating glycoprotein fusogenicity. However, the roles of these two domains are poorly understood. We report here that point deletions and insertions within the MPER or MSD result in functionally inactive proteins. However, when the C-terminal tail domain (CTD) is deleted, the majority of the proteins remain functional. The only residues that were found to be critical for function regardless of the CTD were four hydroxyl-containing amino acids located at the C terminus of the MPER (T139 and T140) and at the N terminus of the MSD (S143 and T144) and a proline near the beginning of the MSD (P148). We demonstrate that hydrogen-bonding at positions T139 and S143 is critical for protein function. Our findings provide novel insights into the role of the MPER in regulating fusogenic activity of viral glycoproteins.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Sequence Determinants in Gammaretroviral Env Cytoplasmic Tails Dictate Virus-Specific Pseudotyping Compatibility.J Virol. 2019 May 15;93(11):e02172-18. doi: 10.1128/JVI.02172-18. Print 2019 Jun 1. J Virol. 2019. PMID: 30894464 Free PMC article.
-
Retrovirus glycoprotein functionality requires proper alignment of the ectodomain and the membrane-proximal cytoplasmic tail.J Virol. 2013 Dec;87(23):12805-13. doi: 10.1128/JVI.01847-13. Epub 2013 Sep 18. J Virol. 2013. PMID: 24049172 Free PMC article.
-
In Vivo Analysis of Infectivity, Fusogenicity, and Incorporation of a Mutagenic Viral Glycoprotein Library Reveals Determinants for Virus Incorporation.J Virol. 2016 Jun 24;90(14):6502-14. doi: 10.1128/JVI.00804-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27147747 Free PMC article.
-
Importance of the short cytoplasmic domain of the feline immunodeficiency virus transmembrane glycoprotein for fusion activity and envelope glycoprotein incorporation into virions.Virology. 2007 Sep 30;366(2):405-14. doi: 10.1016/j.virol.2007.05.019. Epub 2007 Jun 7. Virology. 2007. PMID: 17559903
-
Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection.J Virol. 2008 Jun;82(11):5417-28. doi: 10.1128/JVI.02666-07. Epub 2008 Mar 19. J Virol. 2008. PMID: 18353944 Free PMC article.
Cited by
-
Characterizing the Lassa Virus Envelope Glycoprotein Membrane Proximal External Region for Its Role in Fusogenicity.Virol Sin. 2021 Apr;36(2):273-280. doi: 10.1007/s12250-020-00286-3. Epub 2020 Sep 8. Virol Sin. 2021. PMID: 32897505 Free PMC article.
-
Inhibition of ATM-directed antiviral responses by HIV-1 Vif.PLoS Pathog. 2023 Sep 5;19(9):e1011634. doi: 10.1371/journal.ppat.1011634. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37669285 Free PMC article.
-
Epstein-Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity.Nat Microbiol. 2019 Jan;4(1):78-88. doi: 10.1038/s41564-018-0284-6. Epub 2018 Nov 12. Nat Microbiol. 2019. PMID: 30420783 Free PMC article.
-
Sequence Determinants in Gammaretroviral Env Cytoplasmic Tails Dictate Virus-Specific Pseudotyping Compatibility.J Virol. 2019 May 15;93(11):e02172-18. doi: 10.1128/JVI.02172-18. Print 2019 Jun 1. J Virol. 2019. PMID: 30894464 Free PMC article.
-
Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus.Cell. 2020 Aug 20;182(4):812-827.e19. doi: 10.1016/j.cell.2020.06.043. Epub 2020 Jul 3. Cell. 2020. PMID: 32697968 Free PMC article.
References
-
- Anderson ET, Hayflick LJ, Thomas G. 1993. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed α 1-antitrypsin variant. J Biol Chem 268:. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

