Mismatches in the Influenza A Virus RNA Panhandle Prevent Retinoic Acid-Inducible Gene I (RIG-I) Sensing by Impairing RNA/RIG-I Complex Formation
- PMID: 26446607
- PMCID: PMC4702558
- DOI: 10.1128/JVI.01671-15
Mismatches in the Influenza A Virus RNA Panhandle Prevent Retinoic Acid-Inducible Gene I (RIG-I) Sensing by Impairing RNA/RIG-I Complex Formation
Abstract
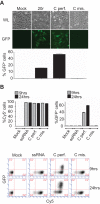
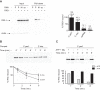
Influenza virus RNA (vRNA) promoter panhandle structures are believed to be sensed by retinoic acid-inducible gene I (RIG-I). The occurrence of mismatches in this double-stranded RNA structure raises questions about their effect on innate sensing. Our results suggest that mismatches in vRNA promoters decrease binding to RIG-I in vivo, affecting RNA/RIG-I complex formation and preventing RIG-I activation. These results can be inferred to apply to other viruses and suggest that mismatches may represent a general viral strategy to escape RIG-I sensing.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514:372–375. doi:10.1038/nature13590. - DOI - PMC - PubMed
-
- Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. doi:10.1016/j.chom.2013.01.012. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

