Distinct Acute Lymphoblastic Leukemia (ALL)-associated Janus Kinase 3 (JAK3) Mutants Exhibit Different Cytokine-Receptor Requirements and JAK Inhibitor Specificities
- PMID: 26446793
- PMCID: PMC4661414
- DOI: 10.1074/jbc.M115.670224
Distinct Acute Lymphoblastic Leukemia (ALL)-associated Janus Kinase 3 (JAK3) Mutants Exhibit Different Cytokine-Receptor Requirements and JAK Inhibitor Specificities
Abstract
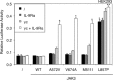
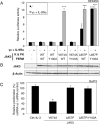
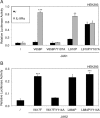
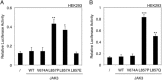
JAK1 and JAK3 are recurrently mutated in acute lymphoblastic leukemia. These tyrosine kinases associate with heterodimeric cytokine receptors such as IL-7 receptor or IL-9 receptor, in which JAK1 is appended to the specific chain, and JAK3 is appended to the common gamma chain. Here, we studied the role of these receptor complexes in mediating the oncogenic activity of JAK3 mutants. Although JAK3(V674A) and the majority of other JAK3 mutants needed to bind to a functional cytokine receptor complex to constitutively activate STAT5, JAK3(L857P) was unexpectedly found to not depend on such receptor complexes for its activity, which was induced without receptor or JAK1 co-expression. Introducing a mutation in the FERM domain that abolished JAK-receptor interaction did not affect JAK3(L857P) activity, whereas it inhibited the other receptor-dependent mutants. The same cytokine receptor independence as for JAK3(L857P) was observed for homologous Leu(857) mutations of JAK1 and JAK2 and for JAK3(L875H). This different cytokine receptor requirement correlated with different functional properties in vivo and with distinct sensitivity to JAK inhibitors. Transduction of murine hematopoietic cells with JAK3(V674A) led homogenously to lymphoblastic leukemias in BALB/c mice. In contrast, transduction with JAK3(L857P) induced various types of lymphoid and myeloid leukemias. Moreover, ruxolitinib, which preferentially blocks JAK1 and JAK2, abolished the proliferation of cells transformed by the receptor-dependent JAK3(V674A), yet proved much less potent on cells expressing JAK3(L857P). These particular cells were, in contrast, more sensitive to JAK3-specific inhibitors. Altogether, our results showed that different JAK3 mutations induce constitutive activation through distinct mechanisms, pointing to specific therapeutic perspectives.
Keywords: JAK inhibitor; Janus kinase (JAK); leukemia; oncogene; signal transduction; tyrosine-protein kinase (tyrosine kinase).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Haan C., Rolvering C., Raulf F., Kapp M., Drückes P., Thoma G., Behrmann I., and Zerwes H. G. (2011) Jak1 has a dominant role over Jak3 in signal transduction through γc-containing cytokine receptors. Chem. Biol. 18, 314–323 - PubMed
-
- Saharinen P., and Silvennoinen O. (2002) The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 277, 47954–47963 - PubMed
-
- Haan C., Is'harc H., Hermanns H. M., Schmitz-Van De Leur H., Kerr I. M., Heinrich P. C., Grötzinger J., and Behrmann I. (2001) Mapping of a region within the N terminus of Jak1 involved in cytokine receptor interaction. J. Biol. Chem. 276, 37451–37458 - PubMed
-
- Tang W., Huo H., Zhu J., Ji H., Zou W., Xu L., Sun L., Zheng Z., Theze J., and Liu X. (2001) Critical sites for the interaction between IL-2Rγ and JAK3 and the following signaling. Biochem. Biophys. Res. Commun. 283, 598–605 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

