Ectopic Expression of the Coleus R2R3 MYB-Type Proanthocyanidin Regulator Gene SsMYB3 Alters the Flower Color in Transgenic Tobacco
- PMID: 26448466
- PMCID: PMC4598174
- DOI: 10.1371/journal.pone.0139392
Ectopic Expression of the Coleus R2R3 MYB-Type Proanthocyanidin Regulator Gene SsMYB3 Alters the Flower Color in Transgenic Tobacco
Abstract
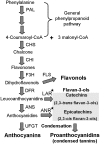
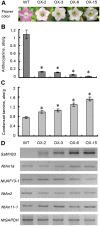
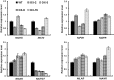
Proanthocyanidins (PAs) play an important role in plant disease defense and have beneficial effects on human health. We isolated and characterized a novel R2R3 MYB-type PA-regulator SsMYB3 from a well-known ornamental plant, coleus (Solenostemon scutellarioides), to study the molecular regulation of PAs and to engineer PAs biosynthesis. The expression level of SsMYB3 was correlated with condensed tannins contents in various coleus tissues and was induced by wounding and light. A complementation test in the Arabidopsis tt2 mutant showed that SsMYB3 could restore the PA-deficient seed coat phenotype and activated expression of the PA-specific gene ANR and two related genes, DFR and ANS. In yeast two-hybrid assays, SsMYB3 interacted with the Arabidopsis AtTT8 and AtTTG1 to reform the ternary transcriptional complex, and also interacted with two tobacco bHLH proteins (NtAn1a and NtJAF13-1) and a WD40 protein, NtAn11-1. Ectopic overexpression of SsMYB3 in transgenic tobacco led to almost-white flowers by greatly reducing anthocyanin levels and enhancing accumulation of condensed tannins. This overexpression of SsMYB3 upregulated the key PA genes (NtLAR and NtANR) and late anthocyanin structural genes (NtDFR and NtANS), but downregulated the expression of the final anthocyanin gene NtUFGT. The formative SsMYB3-complex represses anthocyanin accumulation by directly suppressing the expression of the final anthocyanin structural gene NtUFGT, through competitive inhibition or destabilization of the endogenous NtAn2-complex formation. These results suggested that SsMYB3 may form a transcription activation complex to regulate PA biosynthesis in the Arabidopsis tt2 mutant and transgenic tobacco. Our findings suggest that SsMYB3 is involved in the regulation of PA biosynthesis in coleus and has the potential as a molecular tool for manipulating biosynthesis of PAs in fruits and other crops using metabolic engineering.
Conflict of interest statement
Figures





Similar articles
-
Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco.Planta. 2010 Apr;231(5):1061-76. doi: 10.1007/s00425-010-1108-y. Epub 2010 Feb 16. Planta. 2010. PMID: 20157728
-
A novel R2R3-MYB from grape hyacinth, MaMybA, which is different from MaAN2, confers intense and magenta anthocyanin pigmentation in tobacco.BMC Plant Biol. 2019 Sep 9;19(1):390. doi: 10.1186/s12870-019-1999-0. BMC Plant Biol. 2019. PMID: 31500571 Free PMC article.
-
A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana.Plant Physiol Biochem. 2019 Aug;141:60-72. doi: 10.1016/j.plaphy.2019.05.015. Epub 2019 May 20. Plant Physiol Biochem. 2019. PMID: 31128564
-
Genome-wide identification of Pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in Pistacia chinensis.Int J Biol Macromol. 2021 Dec 1;192:16-27. doi: 10.1016/j.ijbiomac.2021.09.092. Epub 2021 Sep 20. Int J Biol Macromol. 2021. PMID: 34555399 Review.
-
Metabolic engineering of anthocyanins and condensed tannins in plants.Curr Opin Biotechnol. 2013 Apr;24(2):329-35. doi: 10.1016/j.copbio.2012.07.004. Epub 2012 Aug 14. Curr Opin Biotechnol. 2013. PMID: 22901316 Review.
Cited by
-
Comprehensive Influences of Overexpression of a MYB Transcriptor Regulating Anthocyanin Biosynthesis on Transcriptome and Metabolome of Tobacco Leaves.Int J Mol Sci. 2019 Oct 16;20(20):5123. doi: 10.3390/ijms20205123. Int J Mol Sci. 2019. PMID: 31623091 Free PMC article.
-
Nicking Endonuclease-Mediated Vector Construction Strategies for Plant Gene Functional Research.Plants (Basel). 2020 Aug 25;9(9):1090. doi: 10.3390/plants9091090. Plants (Basel). 2020. PMID: 32854250 Free PMC article.
-
In planta high levels of hydrolysable tannins inhibit peroxidase mediated anthocyanin degradation and maintain abaxially red leaves of Excoecaria Cochinchinensis.BMC Plant Biol. 2019 Jul 15;19(1):315. doi: 10.1186/s12870-019-1903-y. BMC Plant Biol. 2019. PMID: 31307378 Free PMC article.
-
Enhancing Flower Color through Simultaneous Expression of the B-peru and mPAP1 Transcription Factors under Control of a Flower-Specific Promoter.Int J Mol Sci. 2018 Jan 20;19(1):309. doi: 10.3390/ijms19010309. Int J Mol Sci. 2018. PMID: 29361688 Free PMC article.
-
AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)-another strategy to modulate pigmentation.Hortic Res. 2019 Jan 1;6:14. doi: 10.1038/s41438-018-0102-6. eCollection 2019. Hortic Res. 2019. PMID: 30603098 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

