Immunological biomarkers predict HIV-1 viral rebound after treatment interruption
- PMID: 26449164
- PMCID: PMC4633715
- DOI: 10.1038/ncomms9495
Immunological biomarkers predict HIV-1 viral rebound after treatment interruption
Abstract
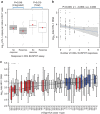
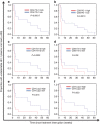
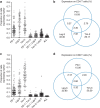
Treatment of HIV-1 infection with antiretroviral therapy (ART) in the weeks following transmission may induce a state of 'post-treatment control' (PTC) in some patients, in whom viraemia remains undetectable when ART is stopped. Explaining PTC could help our understanding of the processes that maintain viral persistence. Here we show that immunological biomarkers can predict time to viral rebound after stopping ART by analysing data from a randomized study of primary HIV-1 infection incorporating a treatment interruption (TI) after 48 weeks of ART (the SPARTAC trial). T-cell exhaustion markers PD-1, Tim-3 and Lag-3 measured prior to ART strongly predict time to the return of viraemia. These data indicate that T-cell exhaustion markers may identify those latently infected cells with a higher proclivity to viral transcription. Our results may open new avenues for understanding the mechanisms underlying PTC, and eventually HIV-1 eradication.
Figures

References
-
- Palella F. J. J. et al.. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV outpatient study investigators. N. Engl. J. Med. 338, 853–860 (1998). - PubMed
-
- Finzi D. et al.. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

