Profiling RNA editing in human tissues: towards the inosinome Atlas
- PMID: 26449202
- PMCID: PMC4598827
- DOI: 10.1038/srep14941
Profiling RNA editing in human tissues: towards the inosinome Atlas
Erratum in
-
Corrigendum: Profiling RNA editing in human tissues: towards the inosinome Atlas.Sci Rep. 2016 Feb 8;6:20755. doi: 10.1038/srep20755. Sci Rep. 2016. PMID: 26854421 Free PMC article. No abstract available.
Abstract
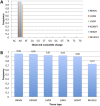
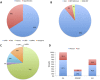
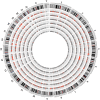
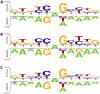
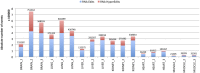
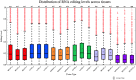
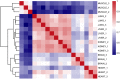
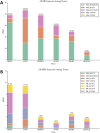
Adenine to Inosine RNA editing is a widespread co- and post-transcriptional mechanism mediated by ADAR enzymes acting on double stranded RNA. It has a plethora of biological effects, appears to be particularly pervasive in humans with respect to other mammals, and is implicated in a number of diverse human pathologies. Here we present the first human inosinome atlas comprising 3,041,422 A-to-I events identified in six tissues from three healthy individuals. Matched directional total-RNA-Seq and whole genome sequence datasets were generated and analysed within a dedicated computational framework, also capable of detecting hyper-edited reads. Inosinome profiles are tissue specific and edited gene sets consistently show enrichment of genes involved in neurological disorders and cancer. Overall frequency of editing also varies, but is strongly correlated with ADAR expression levels. The inosinome database is available at: http://srv00.ibbe.cnr.it/editing/.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

