RNY-derived small RNAs as a signature of coronary artery disease
- PMID: 26449324
- PMCID: PMC4599655
- DOI: 10.1186/s12916-015-0489-y
RNY-derived small RNAs as a signature of coronary artery disease
Abstract
Background: Data from next generation sequencing technologies uncovered the existence of many classes of small RNAs. Recent studies reported that small RNAs are released by cells and can be detected in the blood. In this report, we aimed to discover the occurrence of novel circulating small RNAs in coronary artery disease (CAD).
Methods: We used high-throughput sequencing of small RNAs from human and mouse apoptotic primary macrophages, and analyzed the data by empirical Bayes moderated t-statistics to assess differential expression and the Benjamini and Hochberg method to control the false discovery rate. Results were then confirmed by Northern blot and RT-qPCR in foam cells and in two animal models for atherosclerosis, namely ApoE(-/-) and Ldlr(-/-) mouse lines. Quantitative RT-PCR to detect identified small RNAs, the RNY-derived small RNAs, was performed using sera of 263 patients with CAD compared to 514 matched healthy controls; the Student t-test was applied to statistically assess differences. Associations of small RNAs with clinical characteristics and biological markers were tested using Spearman's rank correlations, while multivariate logistic regressions were performed to test the statistical association of small RNA levels with CAD.
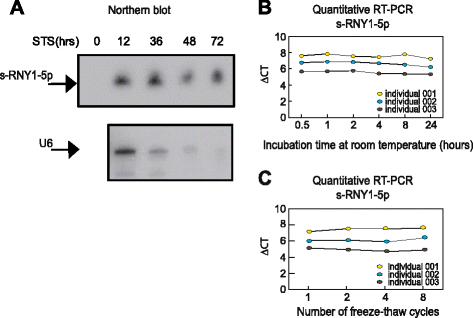
Results: Here, we report that, in macrophages stimulated with pro-apoptotic or pro-atherogenic stimuli, the Ro-associated non-coding RNAs, called RNYs or Y-RNAs, are processed into small RNAs (~24-34 nt) referred to as small-RNYs (s-RNYs), including s-RNY1-5p processed from RNY1. A significant upregulation of s-RNY expression was found in aortic arches and blood plasma from ApoE(-/-) and Ldlr(-/-) mice and in serum from CAD patients (P <0.001). Biostatistical analysis revealed a positive association of s-RNY1-5p with hs-CRP and ApoB levels; however, no statistical interaction was found between either of these two markers and s-RNY1-5p in relation to the CAD status. Levels of s-RNY1-5p were also independent from statin and fibrate therapies.
Conclusion: Our results position the s-RNY1-5p as a relevant novel independent diagnostic biomarker for atherosclerosis-related diseases. Measurement of circulating s-RNY expression would be a valuable companion diagnostic to monitor foam cell apoptosis during atherosclerosis pathogenesis and to evaluate patient's responsiveness to future therapeutic strategies aiming to attenuate apoptosis in foam cells in advanced atherosclerotic lesions.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

