Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers
- PMID: 26449484
- PMCID: PMC4599317
- DOI: 10.1186/s13073-015-0225-4
Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers
Abstract
Background: The mechanisms by which genetic variants, such as single nucleotide polymorphisms (SNPs), identified in genome-wide association studies act to influence body mass remain unknown for most of these SNPs, which continue to puzzle the scientific community. Recent evidence points to the epigenetic and chromatin states of the genome as having important roles.
Methods: We genotyped 355 healthy young individuals for 52 known obesity-associated SNPs and obtained DNA methylation levels in their blood using the Illumina 450 K BeadChip. Associations between alleles and methylation at proximal cytosine residues were tested using a linear model adjusted for age, sex, weight category, and a proxy for blood cell type counts. For replication in other tissues, we used two open-access datasets (skin fibroblasts, n = 62; four brain regions, n = 121-133) and an additional dataset in subcutaneous and visceral fat (n = 149).
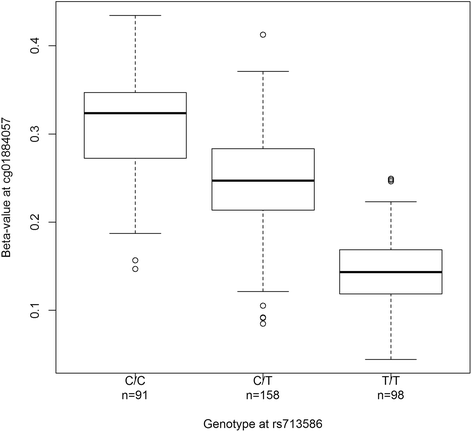
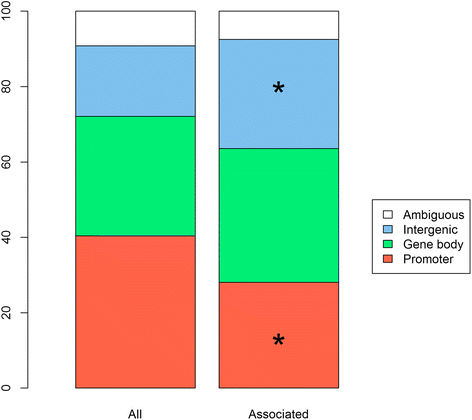
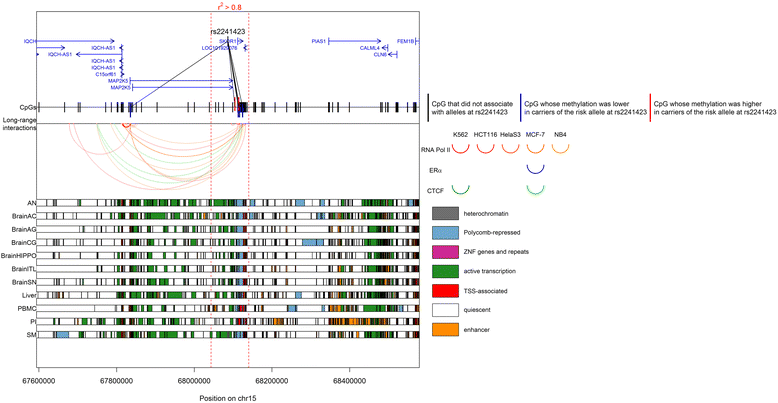
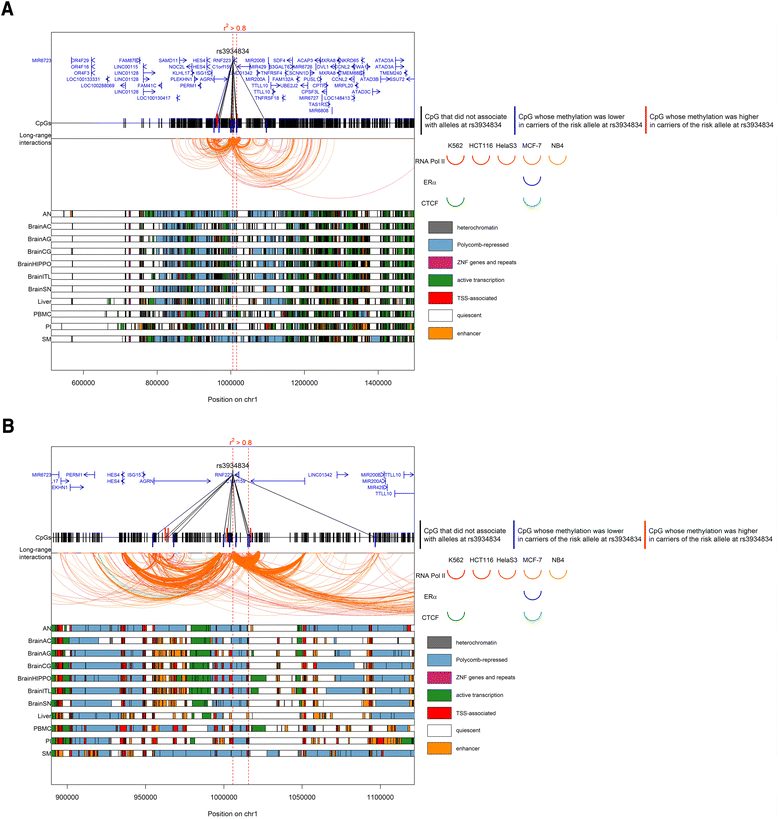
Results: We found that alleles at 28 of these obesity-associated SNPs associate with methylation levels at 107 proximal CpG sites. Out of 107 CpG sites, 38 are located in gene promoters, including genes strongly implicated in obesity (MIR148A, BDNF, PTPMT1, NR1H3, MGAT1, SCGB3A1, HOXC12, PMAIP1, PSIP1, RPS10-NUDT3, RPS10, SKOR1, MAP2K5, SIX5, AGRN, IMMP1L, ELP4, ITIH4, SEMA3G, POMC, ADCY3, SSPN, LGR4, TUFM, MIR4721, SULT1A1, SULT1A2, APOBR, CLN3, SPNS1, SH2B1, ATXN2L, and IL27). Interestingly, the associated SNPs are in known eQTLs for some of these genes. We also found that the 107 CpGs are enriched in enhancers in peripheral blood mononuclear cells. Finally, our results indicate that some of these associations are not blood-specific as we successfully replicated four associations in skin fibroblasts.
Conclusions: Our results strongly suggest that many obesity-associated SNPs are associated with proximal gene regulation, which was reflected by association of obesity risk allele genotypes with differential DNA methylation. This study highlights the importance of DNA methylation and other chromatin marks as a way to understand the molecular basis of genetic variants associated with human diseases and traits.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

