Regulation of IL-8 gene expression in gliomas by microRNA miR-93
- PMID: 26449498
- PMCID: PMC4598972
- DOI: 10.1186/s12885-015-1659-1
Regulation of IL-8 gene expression in gliomas by microRNA miR-93
Abstract
Background: Different strategies have been proposed to target neoangiogenesis in gliomas, besides those targeting Vascular Endothelial Growth Factor (VEGF). The chemokine Interleukin-8 (IL-8) has been shown to possess both tumorigenic and proangiogenic properties. Although different pathways of induction of IL-8 gene expression have been already elucidated, few data are available on its post-transcriptional regulation in gliomas.
Methods: Here we investigated the role of the microRNA miR-93 on the expression levels of IL-8 and other pro-inflammatory genes by RT-qPCR and Bio-Plex analysis. We used different disease model systems, including clinical samples from glioma patients and two glioma cell lines, U251 and T98G.
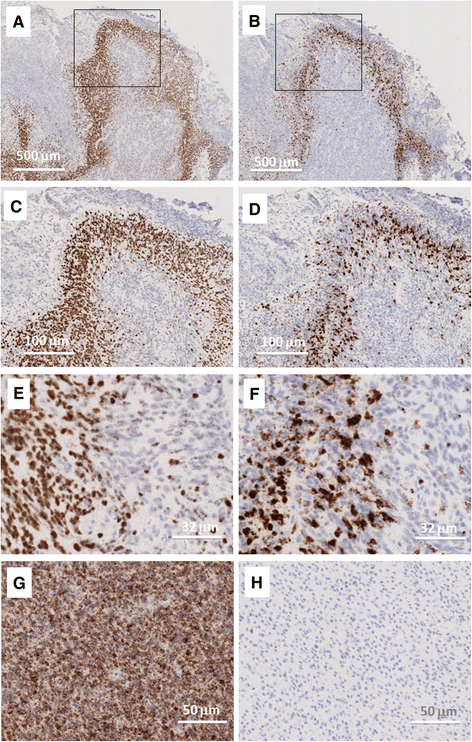
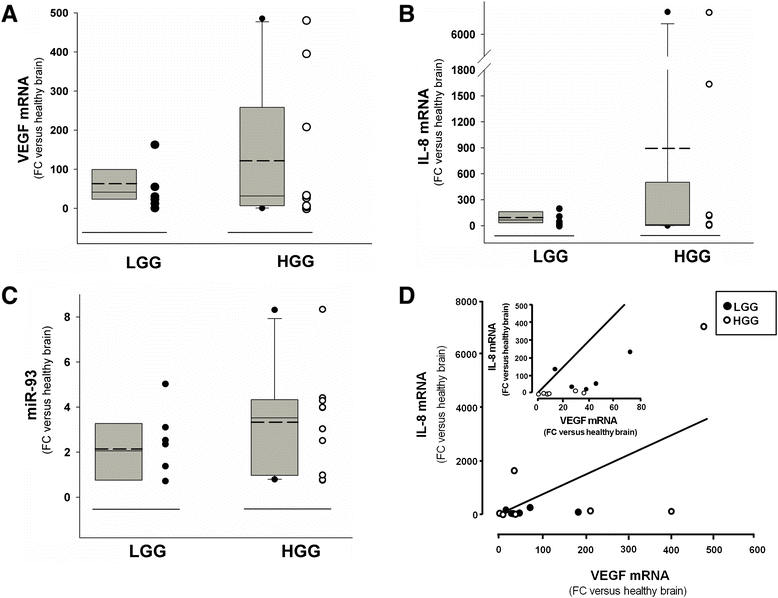
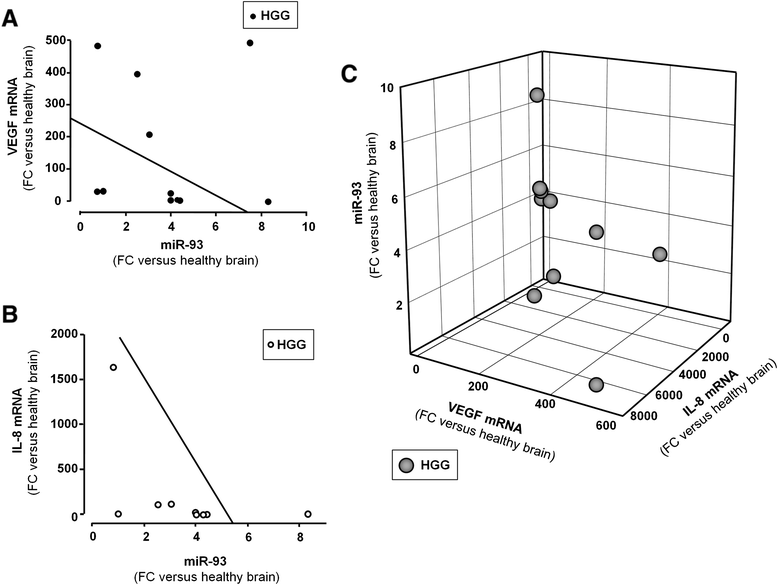
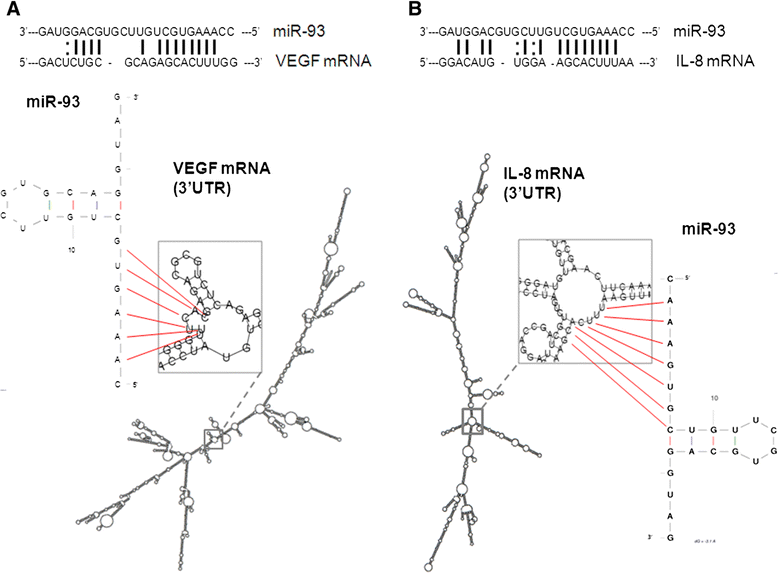
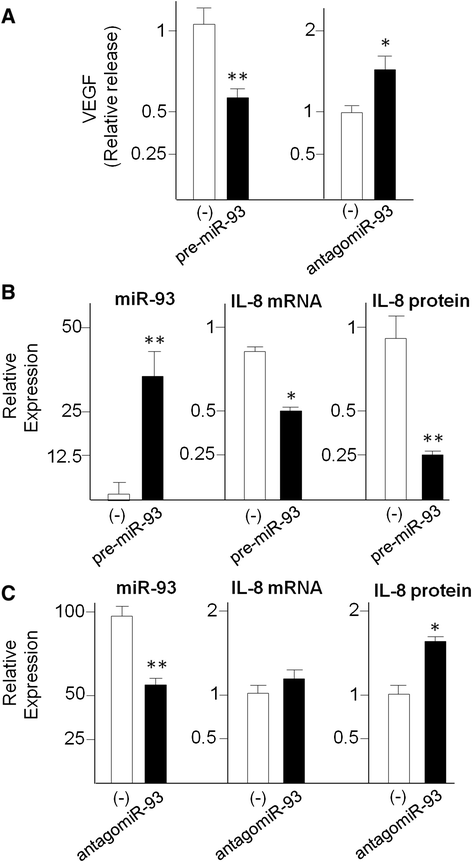
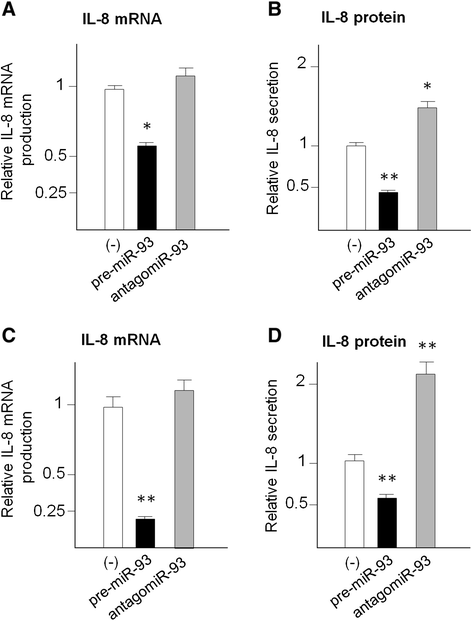
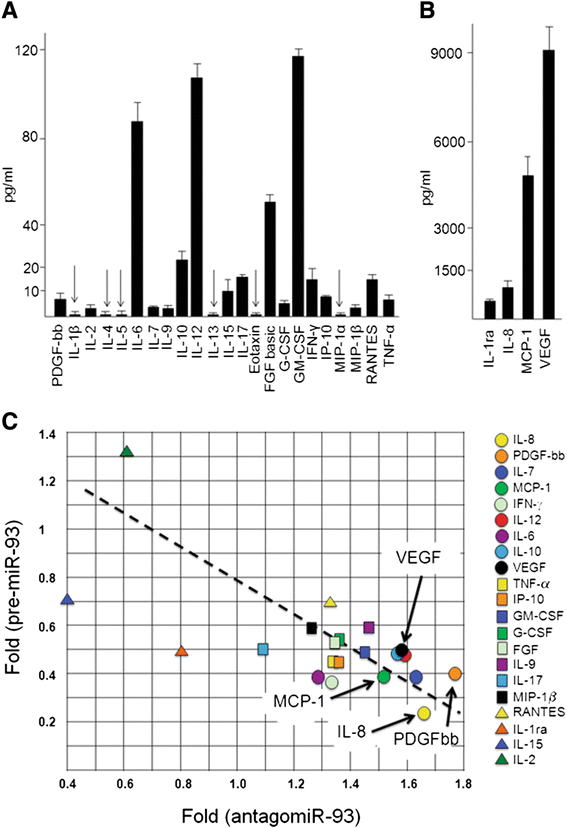
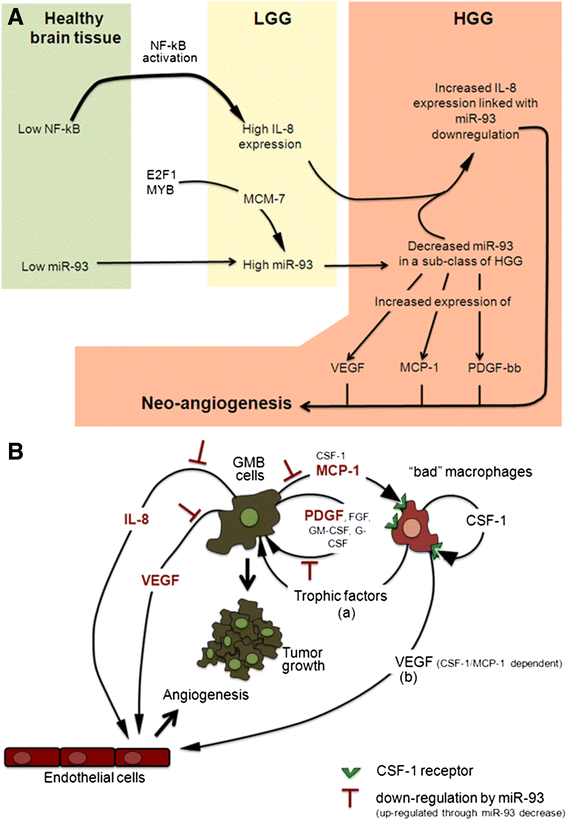
Results: IL-8 and VEGF transcripts are highly expressed in low and high grade gliomas in respect to reference healthy brain; miR-93 expression is also increased and inversely correlated with transcription of IL-8 and VEGF genes. Computational analysis showed the presence of miR-93 consensus sequences in the 3'UTR region of both VEGF and IL-8 mRNAs, predicting possible interaction with miR-93 and suggesting a potential regulatory role of this microRNA. In vitro transfection with pre-miR-93 and antagomiR-93 inversely modulated VEGF and IL-8 gene expression and protein release when the glioma cell line U251 was considered. Similar data were obtained on IL-8 gene regulation in the other glioma cell line analyzed, T98G. The effect of pre-miR-93 and antagomiR-93 in U251 cells has been extended to the secretion of a panel of cytokines, chemokines and growth factors, which consolidated the concept of a role of miR-93 in IL-8 and VEGF gene expression and evidenced a potential regulatory role also for MCP-1 and PDGF (also involved in angiogenesis).
Conclusion: In conclusion, our results suggest an increasing role of miR-93 in regulating the level of expression of several genes involved in the angiogenesis of gliomas.
Figures
Similar articles
-
MicroRNA miR-93-5p regulates expression of IL-8 and VEGF in neuroblastoma SK-N-AS cells.Oncol Rep. 2016 May;35(5):2866-72. doi: 10.3892/or.2016.4676. Epub 2016 Mar 15. Oncol Rep. 2016. PMID: 26986724
-
MicroRNA-7 directly targets insulin-like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas.Diagn Pathol. 2014 Nov 14;9:211. doi: 10.1186/s13000-014-0211-y. Diagn Pathol. 2014. PMID: 25394492 Free PMC article.
-
Reduced expression of microRNA-497 is associated with greater angiogenesis and poor prognosis in human gliomas.Hum Pathol. 2016 Dec;58:47-53. doi: 10.1016/j.humpath.2016.04.022. Epub 2016 Aug 25. Hum Pathol. 2016. PMID: 27569294
-
MicroRNAs as efficient biomarkers in high-grade gliomas.Folia Neuropathol. 2016;54(4):369-374. doi: 10.5114/fn.2016.64812. Folia Neuropathol. 2016. PMID: 28139816 Review.
-
miRNA Regulation in Gliomas: Usual Suspects in Glial Tumorigenesis and Evolving Clinical Applications.J Neuropathol Exp Neurol. 2017 Apr 1;76(4):246-254. doi: 10.1093/jnen/nlx005. J Neuropathol Exp Neurol. 2017. PMID: 28431179 Review.
Cited by
-
Upregulation of miR-93 and inhibition of LIMK1 improve ventricular remodeling and alleviate cardiac dysfunction in rats with chronic heart failure by inhibiting RhoA/ROCK signaling pathway activation.Aging (Albany NY). 2019 Sep 20;11(18):7570-7586. doi: 10.18632/aging.102272. Epub 2019 Sep 20. Aging (Albany NY). 2019. PMID: 31541994 Free PMC article.
-
In vitro induction of interleukin-8 by SARS-CoV-2 Spike protein is inhibited in bronchial epithelial IB3-1 cells by a miR-93-5p agomiR.Int Immunopharmacol. 2021 Dec;101(Pt B):108201. doi: 10.1016/j.intimp.2021.108201. Epub 2021 Sep 28. Int Immunopharmacol. 2021. PMID: 34653729 Free PMC article.
-
Evaluation of Changes in Some Functional Properties of Human Mesenchymal Stromal Cells Induced by Low Doses of Ionizing Radiation.Int J Mol Sci. 2023 Mar 28;24(7):6346. doi: 10.3390/ijms24076346. Int J Mol Sci. 2023. PMID: 37047317 Free PMC article.
-
Effect of Necrosis on the miRNA-mRNA Regulatory Network in CRT-MG Human Astroglioma Cells.Cancer Res Treat. 2018 Apr;50(2):382-397. doi: 10.4143/crt.2016.551. Epub 2017 May 22. Cancer Res Treat. 2018. PMID: 28546527 Free PMC article.
-
Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide.Nucleic Acids Res. 2018 Feb 16;46(3):1424-1440. doi: 10.1093/nar/gkx1221. Nucleic Acids Res. 2018. PMID: 29202181 Free PMC article.
References
-
- Bhattacharya D, Singh MK, Chaudhuri S, Acharya S, Basu AK, Chaudhuri S. T11TS impedes glioma angiogenesis by inhibiting VEGF signaling and pro-survival PI3K/Akt/eNOS pathway with concomitant upregulation of PTEN in brain endothelial cells. J Neurooncol. 2013;113:13–25. doi: 10.1007/s11060-013-1095-5. - DOI - PubMed
-
- Qiu B, Zhang D, Wang Y, Ou S, Wang J, Tao J, et al. Interleukin-6 is overexpressed and augments invasiveness of human glioma stem cells in vitro. Clin Exp Metastasis. 2013;30(8):1009–18. Epub Jul 6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

