A comparative genomics study on the effect of individual amino acids on ribosome stalling
- PMID: 26449596
- PMCID: PMC4602185
- DOI: 10.1186/1471-2164-16-S10-S5
A comparative genomics study on the effect of individual amino acids on ribosome stalling
Abstract
Background: During protein synthesis, the nascent peptide chain emerges from the ribosome through the ribosomal exit tunnel. Biochemical interactions between the nascent peptide and the tunnel may stall the ribosome movement and thus affect the expression level of the protein being synthesized. Earlier studies focused on one model organism (S. cerevisiae), have suggested that certain amino acid sequences may be responsible for ribosome stalling; however, the stalling effect at the individual amino acid level across many organisms has not yet been quantified.
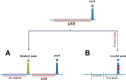
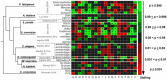
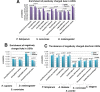
Results: By analyzing multiple ribosome profiling datasets from different organisms (including prokaryotes and eukaryotes), we report for the first time the organism-specific amino acids that significantly lead to ribosome stalling. We show that the identity of the stalling amino acids vary across the tree of life. In agreement with previous studies, we observed a remarkable stalling signal of proline and arginine in S. cerevisiae. In addition, our analysis supports the conjecture that the stalling effect of positively charged amino acids is not universal and that in certain conditions, negative charge may also induce ribosome stalling. Finally, we show that the beginning part of the tunnel tends to undergo more interactions with the translated amino acids than other positions along the tunnel.
Conclusions: The reported results support the conjecture that the ribosomal exit tunnel interacts with various amino acids and that the nature of these interactions varies among different organisms. Our findings should contribute towards better understanding of transcript and proteomic evolution and translation elongation regulation.
Figures


References
-
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. Journal of molecular evolution. 2001;53(4-5):290–298. - PubMed
-
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends in biotechnology. 2004;22(7):346–353. - PubMed
-
- Sørensen MA, Kurland C, Pedersen S. Codon usage determines translation rate in Escherichia coli. Journal of molecular biology. 1989;207(2):365–377. - PubMed
-
- Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process: effect of tRNA availability on the rate of elongation of nascent polypeptide chains. Journal of molecular biology. 1984;180(3):549–576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

