CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice
- PMID: 26449883
- PMCID: PMC4786912
- DOI: 10.1038/mt.2015.192
CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice
Abstract
Duchenne muscular dystrophy (DMD) is a degenerative muscle disease caused by genetic mutations that lead to the disruption of dystrophin in muscle fibers. There is no curative treatment for this devastating disease. Clustered regularly interspaced short palindromic repeat/Cas9 (CRISPR/Cas9) has emerged as a powerful tool for genetic manipulation and potential therapy. Here we demonstrate that CRIPSR-mediated genome editing efficiently excised a 23-kb genomic region on the X-chromosome covering the mutant exon 23 in a mouse model of DMD, and restored dystrophin expression and the dystrophin-glycoprotein complex at the sarcolemma of skeletal muscles in live mdx mice. Electroporation-mediated transfection of the Cas9/gRNA constructs in the skeletal muscles of mdx mice normalized the calcium sparks in response to osmotic shock. Adenovirus-mediated transduction of Cas9/gRNA greatly reduced the Evans blue dye uptake of skeletal muscles at rest and after downhill treadmill running. This study provides proof evidence for permanent gene correction in DMD.
Figures
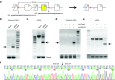
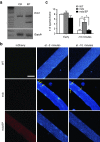
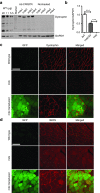
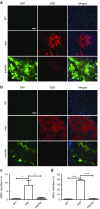
Comment in
-
CRISPR/Cas9 Flexes Its Muscles: In Vivo Somatic Gene Editing for Muscular Dystrophy.Mol Ther. 2016 Mar;24(3):414-6. doi: 10.1038/mt.2016.29. Mol Ther. 2016. PMID: 26952918 Free PMC article. No abstract available.
References
-
- Cohn, RD and Campbell, KP (2000). Molecular basis of muscular dystrophies. Muscle Nerve 23: 1456–1471. - PubMed
-
- Wallace, GQ and McNally, EM (2009). Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol 71: 37–57. - PubMed
-
- Koenig, M, Hoffman, EP, Bertelson, CJ, Monaco, AP, Feener, C and Kunkel, LM (1987). Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50: 509–517. - PubMed
-
- Emery, AE (1991). Population frequencies of inherited neuromuscular diseases–a world survey. Neuromuscul Disord 1: 19–29. - PubMed
-
- Campbell, KP (1995). Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell 80: 675–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

