Structure of the DBL3X-DBL4ε region of the VAR2CSA placental malaria vaccine candidate: insight into DBL domain interactions
- PMID: 26450557
- PMCID: PMC4598876
- DOI: 10.1038/srep14868
Structure of the DBL3X-DBL4ε region of the VAR2CSA placental malaria vaccine candidate: insight into DBL domain interactions
Abstract
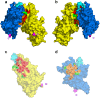
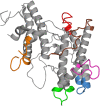
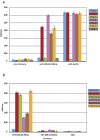
The human malaria parasite, Plasmodium falciparum, is able to evade spleen-mediated clearing from blood stream by sequestering in peripheral organs. This is due to the adhesive properties conferred by the P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1) family exported by the parasite to the surface of infected erythrocytes. Expression of the VAR2CSA variant of PfEMP1 leads to pregnancy-associated malaria, which occurs when infected erythrocytes massively sequester in the placenta by binding to low-sulfated Chondroitin Sulfate A (CSA) present in the intervillous spaces. VAR2CSA is a 350 kDa protein that carries six Duffy-Binding Like (DBL) domains, one Cysteine-rich Inter-Domain Regions (CIDR) and several inter-domain regions. In the present paper, we report for the first time the crystal structure at 2.9 Å of a VAR2CSA double domain, DBL3X-DBL4ε, from the FCR3 strain. DBL3X and DBL4ε share a large contact interface formed by residues that are invariant or highly conserved in VAR2CSA variants, which suggests that these two central DBL domains (DBL3X-DBL4ε) contribute significantly to the structuring of the functional VAR2CSA extracellular region. We have also examined the antigenicity of peptides corresponding to exposed loop regions of the DBL4ε structure.
Figures

Similar articles
-
Structural and immunological correlations between the variable blocks of the VAR2CSA domain DBL6ε from two Plasmodium falciparum parasite lines.J Mol Biol. 2013 May 27;425(10):1697-711. doi: 10.1016/j.jmb.2013.02.014. Epub 2013 Feb 18. J Mol Biol. 2013. PMID: 23429057
-
Antibodies to Escherichia coli-expressed C-terminal domains of Plasmodium falciparum variant surface antigen 2-chondroitin sulfate A (VAR2CSA) inhibit binding of CSA-adherent parasites to placental tissue.Infect Immun. 2013 Apr;81(4):1031-9. doi: 10.1128/IAI.00978-12. Epub 2013 Jan 14. Infect Immun. 2013. PMID: 23319559 Free PMC article.
-
Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria.J Biol Chem. 2012 Jul 6;287(28):23332-45. doi: 10.1074/jbc.M112.348839. Epub 2012 May 8. J Biol Chem. 2012. PMID: 22570492 Free PMC article.
-
Designing a VAR2CSA-based vaccine to prevent placental malaria.Vaccine. 2015 Dec 22;33(52):7483-8. doi: 10.1016/j.vaccine.2015.10.011. Epub 2015 Nov 26. Vaccine. 2015. PMID: 26469717 Free PMC article. Review.
-
Progress and Insights Toward an Effective Placental Malaria Vaccine.Front Immunol. 2021 Feb 25;12:634508. doi: 10.3389/fimmu.2021.634508. eCollection 2021. Front Immunol. 2021. PMID: 33717176 Free PMC article. Review.
Cited by
-
Molecular architecture and domain arrangement of the placental malaria protein VAR2CSA suggests a model for carbohydrate binding.J Biol Chem. 2020 Dec 25;295(52):18589-18603. doi: 10.1074/jbc.RA120.014676. Epub 2020 Oct 29. J Biol Chem. 2020. PMID: 33122198 Free PMC article.
-
Placental malaria vaccine candidate antigen VAR2CSA displays atypical domain architecture in some Plasmodium falciparum strains.Commun Biol. 2019 Dec 6;2:457. doi: 10.1038/s42003-019-0704-z. eCollection 2019. Commun Biol. 2019. PMID: 31840102 Free PMC article.
-
Novel Strategies for Malaria Vaccine Design.Front Immunol. 2018 Nov 29;9:2769. doi: 10.3389/fimmu.2018.02769. eCollection 2018. Front Immunol. 2018. PMID: 30555463 Free PMC article. Review.
-
Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.Nat Commun. 2021 May 19;12(1):2956. doi: 10.1038/s41467-021-23254-1. Nat Commun. 2021. PMID: 34011972 Free PMC article.
-
Malaria Parasites: The Great Escape.Front Immunol. 2016 Nov 7;7:463. doi: 10.3389/fimmu.2016.00463. eCollection 2016. Front Immunol. 2016. PMID: 27872623 Free PMC article. Review.
References
-
- Smith J. D., Subramanian G., Gamain B., Baruch D. I. & Miller L. H. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol 110, 293–310 (2000). - PubMed
-
- Fried M. & Duffy P. E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272, 1502–1504 (1996). - PubMed
-
- Salanti A. et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49, 179–191 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

