Epigenetic Basis of Mental Illness
- PMID: 26450593
- PMCID: PMC4826318
- DOI: 10.1177/1073858415608147
Epigenetic Basis of Mental Illness
Abstract
Psychiatric disorders are complex multifactorial illnesses involving chronic alterations in neural circuit structure and function as well as likely abnormalities in glial cells. While genetic factors are important in the etiology of most mental disorders, the relatively high rates of discordance among identical twins, particularly for depression and other stress-related syndromes, clearly indicate the importance of additional mechanisms. Environmental factors such as stress are known to play a role in the onset of these illnesses. Exposure to such environmental insults induces stable changes in gene expression, neural circuit function, and ultimately behavior, and these maladaptations appear distinct between developmental versus adult exposures. Increasing evidence indicates that these sustained abnormalities are maintained by epigenetic modifications in specific brain regions. Indeed, transcriptional dysregulation and the aberrant epigenetic regulation that underlies this dysregulation is a unifying theme in psychiatric disorders. Here, we provide a progress report of epigenetic studies of the three major psychiatric syndromes, depression, schizophrenia, and bipolar disorder. We review the literature derived from animal models of these disorders as well as from studies of postmortem brain tissue from human patients. While epigenetic studies of mental illness remain at early stages, understanding how environmental factors recruit the epigenetic machinery within specific brain regions to cause lasting changes in disease susceptibility and pathophysiology is revealing new insight into the etiology and treatment of these conditions.
Keywords: 3D chromatin structure; DNA methylation; bipolar disorder; depression; histone acetylation; histone methylation; microRNA; schizophrenia.
© The Author(s) 2015.
Conflict of interest statement
Declaration of Conflicting Interests The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
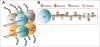
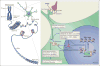

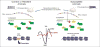

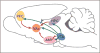
References
-
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:60–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

