Dithiothreitol (DTT) Acts as a Specific, UV-inducible Cross-linker in Elucidation of Protein-RNA Interactions
- PMID: 26450613
- PMCID: PMC4762631
- DOI: 10.1074/mcp.M115.052795
Dithiothreitol (DTT) Acts as a Specific, UV-inducible Cross-linker in Elucidation of Protein-RNA Interactions
Abstract
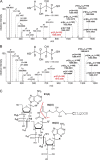
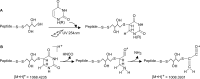
Protein-RNA cross-linking by UV irradiation at 254 nm wavelength has been established as an unbiased method to identify proteins in direct contact with RNA, and has been successfully applied to investigate the spatial arrangement of protein and RNA in large macromolecular assemblies, e.g. ribonucleoprotein-complex particles (RNPs). The mass spectrometric analysis of such peptide-RNA cross-links provides high resolution structural data to the point of mapping protein-RNA interactions to specific peptides or even amino acids. However, the approach suffers from the low yield of cross-linking products, which can be addressed by improving enrichment and analysis methods. In the present article, we introduce dithiothreitol (DTT) as a potent protein-RNA cross-linker. In order to evaluate the efficiency and specificity of DTT, we used two systems, a small synthetic peptide from smB protein incubated with U1 snRNA oligonucleotide and native ribonucleoprotein complexes from S. cerevisiae. Our results unambiguously show that DTT covalently participates in cysteine-uracil crosslinks, which is observable as a mass increment of 151.9966 Da (C(4)H(8)S(2)O(2)) upon mass spectrometric analysis. DTT presents advantages for cross-linking of cysteine containing regions of proteins. This is evidenced by comparison to experiments where (tris(2-carboxyethyl)phosphine) is used as reducing agent, and significantly less cross-links encompassing cysteine residues are found. We further propose insertion of DTT between the cysteine and uracil reactive sites as the most probable structure of the cross-linking products.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 - PubMed
-
- Petrotchenko E. V., and Borchers C. H. (2010) Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 29, 862–876 - PubMed
-
- Paramelle D., Miralles G., Subra G., and Martinez J. (2013) Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics 13, 438–456 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

