Dual Roles of RNF2 in Melanoma Progression
- PMID: 26450788
- PMCID: PMC4670809
- DOI: 10.1158/2159-8290.CD-15-0493
Dual Roles of RNF2 in Melanoma Progression
Abstract
Epigenetic regulators have emerged as critical factors governing the biology of cancer. Here, in the context of melanoma, we show that RNF2 is prognostic, exhibiting progression-correlated expression in human melanocytic neoplasms. Through a series of complementary gain-of-function and loss-of-function studies in mouse and human systems, we establish that RNF2 is oncogenic and prometastatic. Mechanistically, RNF2-mediated invasive behavior is dependent on its ability to monoubiquitinate H2AK119 at the promoter of LTBP2, resulting in silencing of this negative regulator of TGFβ signaling. In contrast, RNF2's oncogenic activity does not require its catalytic activity nor does it derive from its canonical gene repression function. Instead, RNF2 drives proliferation through direct transcriptional upregulation of the cell-cycle regulator CCND2. We further show that MEK1-mediated phosphorylation of RNF2 promotes recruitment of activating histone modifiers UTX and p300 to a subset of poised promoters, which activates gene expression. In summary, RNF2 regulates distinct biologic processes in the genesis and progression of melanoma via different molecular mechanisms.
Significance: The role of epigenetic regulators in cancer progression is being increasingly appreciated. We show novel roles for RNF2 in melanoma tumorigenesis and metastasis, albeit via different mechanisms. Our findings support the notion that epigenetic regulators, such as RNF2, directly and functionally control powerful gene networks that are vital in multiple cancer processes.
©2015 American Association for Cancer Research.
Figures
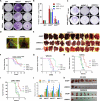
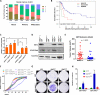
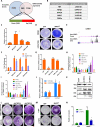
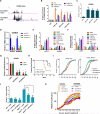

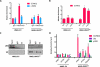
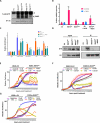
Comment in
-
RNF2 E3 or Not to E3: Dual Roles of RNF2 Overexpression in Melanoma.Cancer Discov. 2015 Dec;5(12):1241-3. doi: 10.1158/2159-8290.CD-15-1285. Cancer Discov. 2015. PMID: 26637658 Free PMC article.
Similar articles
-
Down-regulation of salt-inducible kinase 1 (SIK1) is mediated by RNF2 in hepatocarcinogenesis.Oncotarget. 2017 Jan 10;8(2):3144-3155. doi: 10.18632/oncotarget.13673. Oncotarget. 2017. PMID: 27911266 Free PMC article.
-
Ring finger protein 2 promotes colorectal cancer progression by suppressing early growth response 1.Aging (Albany NY). 2020 Dec 19;12(24):26199-26220. doi: 10.18632/aging.202396. Epub 2020 Dec 19. Aging (Albany NY). 2020. PMID: 33346749 Free PMC article.
-
Radiosensitization of esophageal carcinoma cells by knockdown of RNF2 expression.Int J Oncol. 2016 May;48(5):1985-96. doi: 10.3892/ijo.2016.3404. Epub 2016 Feb 19. Int J Oncol. 2016. PMID: 26936624
-
Repression of TGF-beta signaling by the oncogenic protein SKI in human melanomas: consequences for proliferation, survival, and metastasis.Oncogene. 2003 May 19;22(20):3123-9. doi: 10.1038/sj.onc.1206452. Oncogene. 2003. PMID: 12793438 Review.
-
Emerging role of RNF2 in cancer: From bench to bedside.J Cell Physiol. 2021 Aug;236(8):5453-5465. doi: 10.1002/jcp.30260. Epub 2021 Jan 5. J Cell Physiol. 2021. PMID: 33400276 Review.
Cited by
-
The histone demethylase PHF8 regulates TGFβ signaling and promotes melanoma metastasis.Sci Adv. 2022 Feb 18;8(7):eabi7127. doi: 10.1126/sciadv.abi7127. Epub 2022 Feb 18. Sci Adv. 2022. PMID: 35179962 Free PMC article.
-
E3 Ubiquitin Ligase Ring Finger Protein 2 Alleviates Cerebral Ischemia-Reperfusion Injury by Stabilizing Mesencephalic Astrocyte-Derived Neurotrophic Factor Through Monoubiquitination.CNS Neurosci Ther. 2024 Nov;30(11):e70136. doi: 10.1111/cns.70136. CNS Neurosci Ther. 2024. PMID: 39614674 Free PMC article.
-
Nuclear P38: Roles in Physiological and Pathological Processes and Regulation of Nuclear Translocation.Int J Mol Sci. 2020 Aug 24;21(17):6102. doi: 10.3390/ijms21176102. Int J Mol Sci. 2020. PMID: 32847129 Free PMC article. Review.
-
RING1B contributes to Ewing sarcoma development by repressing the NaV1.6 sodium channel and the NF-κB pathway, independently of the fusion oncoprotein.Oncotarget. 2016 Jul 19;7(29):46283-46300. doi: 10.18632/oncotarget.10092. Oncotarget. 2016. PMID: 27317769 Free PMC article.
-
Molecular determinants of drug response in TNBC cell lines.Breast Cancer Res Treat. 2020 Jan;179(2):337-347. doi: 10.1007/s10549-019-05473-9. Epub 2019 Oct 26. Breast Cancer Res Treat. 2020. PMID: 31655920 Free PMC article.
References
-
- Leong SP, Gershenwald JE, Soong SJ, Schadendorf D, Tarhini AA, Agarwala S, et al. Cutaneous melanoma: a model to study cancer metastasis. Journal of surgical oncology. 2011;103:538–49. - PubMed
-
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–60. - PubMed
-
- Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochemical Society transactions. 2012;40:341–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

