Suppression of epithelial differentiation by Foxi3 is essential for molar crown patterning
- PMID: 26450968
- PMCID: PMC6517835
- DOI: 10.1242/dev.124172
Suppression of epithelial differentiation by Foxi3 is essential for molar crown patterning
Abstract
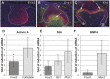
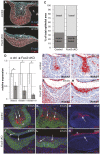
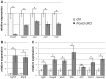
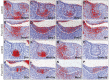
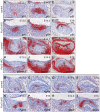
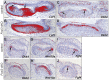
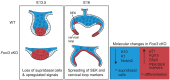
Epithelial morphogenesis generates the shape of the tooth crown. This is driven by patterned differentiation of cells into enamel knots, root-forming cervical loops and enamel-forming ameloblasts. Enamel knots are signaling centers that define the positions of cusp tips in a tooth by instructing the adjacent epithelium to fold and proliferate. Here, we show that the forkhead-box transcription factor Foxi3 inhibits formation of enamel knots and cervical loops and thus the differentiation of dental epithelium in mice. Conditional deletion of Foxi3 (Foxi3 cKO) led to fusion of molars with abnormally patterned shallow cusps. Foxi3 was expressed in the epithelium, and its expression was reduced in the enamel knots and cervical loops and in ameloblasts. Bmp4, a known inducer of enamel knots and dental epithelial differentiation, downregulated Foxi3 in wild-type teeth. Using genome-wide gene expression profiling, we showed that in Foxi3 cKO there was an early upregulation of differentiation markers, such as p21, Fgf15 and Sfrp5. Different signaling pathway components that are normally restricted to the enamel knots were expanded in the epithelium, and Sostdc1, a marker of the intercuspal epithelium, was missing. These findings indicated that the activator-inhibitor balance regulating cusp patterning was disrupted in Foxi3 cKO. In addition, early molar bud morphogenesis and, in particular, formation of the suprabasal epithelial cell layer were impaired. We identified keratin 10 as a marker of suprabasal epithelial cells in teeth. Our results suggest that Foxi3 maintains dental epithelial cells in an undifferentiated state and thereby regulates multiple stages of tooth morphogenesis.
Keywords: Epithelium; Foxi3; Morphogenesis; Multituberculata; Signaling center; Tooth development.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


Similar articles
-
Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation.Adv Dent Res. 2001 Aug;15:14-8. doi: 10.1177/08959374010150010401. Adv Dent Res. 2001. PMID: 12640732 Review.
-
Modulation of activin/bone morphogenetic protein signaling by follistatin is required for the morphogenesis of mouse molar teeth.Dev Dyn. 2004 Sep;231(1):98-108. doi: 10.1002/dvdy.20118. Dev Dyn. 2004. PMID: 15305290
-
Unicuspid and bicuspid tooth crown formation in squamates.J Exp Zool B Mol Dev Evol. 2011 Dec 15;316(8):598-608. doi: 10.1002/jez.b.21438. Epub 2011 Sep 19. J Exp Zool B Mol Dev Evol. 2011. PMID: 21932327
-
Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis.Dev Biol. 2004 Jun 1;270(1):76-93. doi: 10.1016/j.ydbio.2004.02.012. Dev Biol. 2004. PMID: 15136142
-
Reiterative signaling and patterning during mammalian tooth morphogenesis.Mech Dev. 2000 Mar 15;92(1):19-29. doi: 10.1016/s0925-4773(99)00322-6. Mech Dev. 2000. PMID: 10704885 Review.
Cited by
-
The dental phenotype of hairless dogs with FOXI3 haploinsufficiency.Sci Rep. 2017 Jul 14;7(1):5459. doi: 10.1038/s41598-017-05764-5. Sci Rep. 2017. PMID: 28710361 Free PMC article.
-
Clinical presentation and genetic profiles of Chinese patients with velocardiofacial syndrome in a large referral centre.J Genet. 2019 Jun;98(2):42. J Genet. 2019. PMID: 31204702
-
Plasticity within the niche ensures the maintenance of a Sox2+ stem cell population in the mouse incisor.Development. 2018 Jan 8;145(1):dev155929. doi: 10.1242/dev.155929. Development. 2018. PMID: 29180573 Free PMC article.
-
FOXI3 pathogenic variants cause one form of craniofacial microsomia.Nat Commun. 2023 Apr 11;14(1):2026. doi: 10.1038/s41467-023-37703-6. Nat Commun. 2023. PMID: 37041148 Free PMC article.
-
A Hedgehog-Foxf axis coordinates dental follicle-derived alveolar bone formation.Nat Commun. 2025 Jul 2;16(1):6061. doi: 10.1038/s41467-025-61050-3. Nat Commun. 2025. PMID: 40593802 Free PMC article.
References
-
- Andl T., Ahn K., Kairo A., Chu E. Y., Wine-Lee L., Reddy S. T., Croft N. J., Cebra-Thomas J. A., Metzger D., Chambon P. et al. (2004). Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 131, 2257-2268. 10.1242/dev.01125 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

