Pausing on Polyribosomes: Make Way for Elongation in Translational Control
- PMID: 26451481
- PMCID: PMC4600128
- DOI: 10.1016/j.cell.2015.09.041
Pausing on Polyribosomes: Make Way for Elongation in Translational Control
Abstract
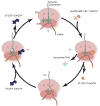
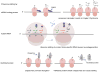
Among the three phases of mRNA translation-initiation, elongation, and termination-initiation has traditionally been considered to be rate limiting and thus the focus of regulation. Emerging evidence, however, demonstrates that control of ribosome translocation (polypeptide elongation) can also be regulatory and indeed exerts a profound influence on development, neurologic disease, and cell stress. The correspondence of mRNA codon usage and the relative abundance of their cognate tRNAs is equally important for mediating the rate of polypeptide elongation. Here, we discuss recent results showing that ribosome pausing is a widely used mechanism for controlling translation and, as a result, biological transitions in health and disease.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

References
-
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. - PubMed
-
- Ballinger DG, Pardue ML. The control of protein synthesis during heat shock in Drosophila cells involves altered polypeptide elongation rates. Cell. 1983;33:103–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

