Event inference in multidomain families with phylogenetic reconciliation
- PMID: 26451642
- PMCID: PMC4610023
- DOI: 10.1186/1471-2105-16-S14-S8
Event inference in multidomain families with phylogenetic reconciliation
Abstract
Background: Reconstructing evolution provides valuable insights into the processes of gene evolution and function. However, while there have been great advances in algorithms and software to reconstruct the history of gene families, these tools do not model the domain shuffling events (domain duplication, insertion, transfer, and deletion) that drive the evolution of multidomain protein families. Protein evolution through domain shuffling events allows for rapid exploration of functions by introducing new combinations of existing folds. This powerful mechanism was key to some significant evolutionary innovations, such as multicellularity and the vertebrate immune system. A method for reconstructing this important evolutionary process is urgently needed.
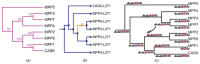
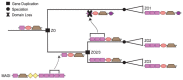
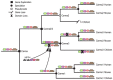
Results: Here, we introduce a novel, event-based framework for studying multidomain evolution by reconciling a domain tree with a gene tree, with additional information provided by the species tree. In the context of this framework, we present the first reconciliation algorithms to infer domain shuffling events, while addressing the challenges inherent in the inference of evolution across three levels of organization.
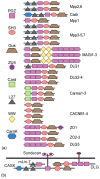
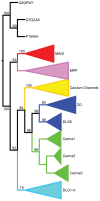
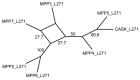
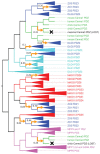
Conclusions: We apply these methods to the evolution of domains in the Membrane associated Guanylate Kinase family. These case studies reveal a more vivid and detailed evolutionary history than previously provided. Our algorithms have been implemented in software, freely available at http://www.cs.cmu.edu/˜durand/Notung.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

