Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore
- PMID: 26451707
- PMCID: PMC5065927
- DOI: 10.1021/acs.analchem.5b03350
Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore
Abstract
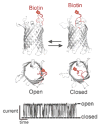
Outer membrane protein G is a monomeric β-barrel porin that has seven flexible loops on its extracellular side. Conformational changes of these labile loops induce gating spikes in current recordings that we exploited as the prime sensing element for protein detection. The gating characteristics, open probability, frequency, and current decrease, provide rich information for analyte identification. Here, we show that two antibiotin antibodies each induced a distinct gating pattern, which allowed them to be readily detected and simultaneously discriminated by a single OmpG nanopore in the presence of fetal bovine serum. Our results demonstrate the feasibility of directly profiling proteins in real-world samples with minimal or no sample pretreatment.
Figures



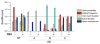
Similar articles
-
Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore.ACS Nano. 2015 Feb 24;9(2):1089-98. doi: 10.1021/nn506606e. Epub 2015 Jan 22. ACS Nano. 2015. PMID: 25575121 Free PMC article.
-
Electrostatic Interactions between OmpG Nanopore and Analyte Protein Surface Can Distinguish between Glycosylated Isoforms.J Phys Chem B. 2015 Aug 13;119(32):10198-206. doi: 10.1021/acs.jpcb.5b06435. Epub 2015 Jul 30. J Phys Chem B. 2015. PMID: 26181080 Free PMC article.
-
Modifying the pH sensitivity of OmpG nanopore for improved detection at acidic pH.Biophys J. 2022 Mar 1;121(5):731-741. doi: 10.1016/j.bpj.2022.01.023. Epub 2022 Feb 5. Biophys J. 2022. PMID: 35131293 Free PMC article.
-
Ion-channels: goals for function-oriented synthesis.Acc Chem Res. 2013 Dec 17;46(12):2773-80. doi: 10.1021/ar400007w. Epub 2013 May 7. Acc Chem Res. 2013. PMID: 23651489 Review.
-
Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers.Chem Phys Lipids. 2006 Jun;141(1-2):30-47. doi: 10.1016/j.chemphyslip.2006.02.004. Epub 2006 Mar 20. Chem Phys Lipids. 2006. PMID: 16581049 Review.
Cited by
-
Programmable nano-reactors for stochastic sensing.Nat Commun. 2021 Oct 4;12(1):5811. doi: 10.1038/s41467-021-26054-9. Nat Commun. 2021. PMID: 34608151 Free PMC article.
-
DNA-assisted oligomerization of pore-forming toxin monomers into precisely-controlled protein channels.Nucleic Acids Res. 2017 Dec 1;45(21):12057-12068. doi: 10.1093/nar/gkx990. Nucleic Acids Res. 2017. PMID: 29088457 Free PMC article.
-
Aberrantly Large Single-Channel Conductance of Polyhistidine Arm-Containing Protein Nanopores.Biochemistry. 2017 Sep 12;56(36):4895-4905. doi: 10.1021/acs.biochem.7b00577. Epub 2017 Aug 28. Biochemistry. 2017. PMID: 28812882 Free PMC article.
-
Sculpting conducting nanopore size and shape through de novo protein design.bioRxiv [Preprint]. 2023 Dec 20:2023.12.20.572500. doi: 10.1101/2023.12.20.572500. bioRxiv. 2023. Update in: Science. 2024 Jul 19;385(6706):282-288. doi: 10.1126/science.adn3796. PMID: 38187764 Free PMC article. Updated. Preprint.
-
PlyAB Nanopores Detect Single Amino Acid Differences in Folded Haemoglobin from Blood.Angew Chem Int Ed Engl. 2022 Aug 22;61(34):e202206227. doi: 10.1002/anie.202206227. Epub 2022 Jul 13. Angew Chem Int Ed Engl. 2022. PMID: 35759385 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

