IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in Arabidopsis
- PMID: 26451844
- PMCID: PMC4599859
- DOI: 10.1371/journal.pgen.1005584
IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in Arabidopsis
Erratum in
-
Correction: IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in Arabidopsis.PLoS Genet. 2016 Jan 6;12(1):e1005795. doi: 10.1371/journal.pgen.1005795. eCollection 2016 Jan. PLoS Genet. 2016. PMID: 26735692 Free PMC article.
Abstract
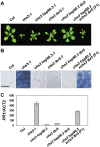
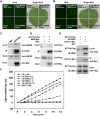
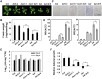
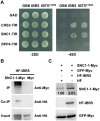
Plant responses to low temperature are tightly associated with defense responses. We previously characterized the chilling-sensitive mutant chs3-1 resulting from the activation of the Toll and interleukin 1 receptor-nucleotide binding-leucine-rich repeat (TIR-NB-LRR)-type resistance (R) protein harboring a C-terminal LIM (Lin-11, Isl-1 and Mec-3 domains) domain. Here we report the identification of a suppressor of chs3, ibr5-7 (indole-3-butyric acid response 5), which largely suppresses chilling-activated defense responses. IBR5 encodes a putative dual-specificity protein phosphatase. The accumulation of CHS3 protein at chilling temperatures is inhibited by the IBR5 mutation. Moreover, chs3-conferred defense phenotypes were synergistically suppressed by mutations in HSP90 and IBR5. Further analysis showed that IBR5, with holdase activity, physically associates with CHS3, HSP90 and SGT1b (Suppressor of the G2 allele of skp1) to form a complex that protects CHS3. In addition to the positive role of IBR5 in regulating CHS3, IBR5 is also involved in defense responses mediated by R genes, including SNC1 (Suppressor of npr1-1, Constitutive 1), RPS4 (Resistance to P. syringae 4) and RPM1 (Resistance to Pseudomonas syringae pv. maculicola 1). Thus, the results of the present study reveal a role for IBR5 in the regulation of multiple R protein-mediated defense responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- de Jong CF, Takken FL, Cai X, de Wit PJ, Joosten MH (2002) Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol Plant Microbe Interact 15: 1040–1049. - PubMed
-
- Xiao S, Charoenwattana P, Holcombe L, Turner JG (2003) The Arabidopsis genes RPW8.1 and RPW8.2 confer induced resistance to powdery mildew diseases in tobacco. Mol Plant Microbe Interact 16: 289–294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

