Thiol versus hydroxamate as zinc binding group in HDAC inhibition: An ab initio QM/MM molecular dynamics study
- PMID: 26452222
- PMCID: PMC4618035
- DOI: 10.1002/jcc.24203
Thiol versus hydroxamate as zinc binding group in HDAC inhibition: An ab initio QM/MM molecular dynamics study
Abstract
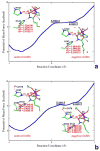
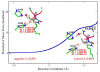
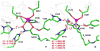
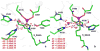
Zinc-dependent histone deacetylases (HDACs) play a critical role in transcriptional repression and gene silencing, and are among the most attractive targets for the development of new therapeutics against cancer and various other diseases. Two HDAC inhibitors have been approved by FDA as anti-cancer drugs: one is SAHA whose hydroxamate is directly bound to zinc, the other is FK228 whose active form may use thiol as the zinc binding group. In spite of extensive studies, it remains to be ambiguous regarding how thiol and hydroxamate are bound to the zinc active site of HDACs. In this work, our computational approaches center on Born-Oppenheimer ab initio quantum mechanical/molecular mechanical (QM/MM) molecular dynamics with umbrella sampling, which allow for modeling of the zinc active site with reasonable accuracy while properly including dynamics and effects of protein environment. Meanwhile, an improved short-long effective function (SLEF2) to describe non-bonded interactions between zinc and other atoms has been employed in initial MM equilibrations. Our ab initio QM/MM MD simulations have confirmed that hydroxamate is neutral when it is bound to HDAC8, and found that thiol is deprotonated when directly bound to zinc in the HDAC active site. By comparing thiol and hydroxamate, our results elucidated the differences in their binding environment in the HDAC active sites, and emphasized the importance of the linker design to achieve more specific binding toward class IIa HDACs.
Keywords: HDAC; QM/MM; Zinc enzyme; inhibitor; molecular dynamics.
© 2015 Wiley Periodicals, Inc.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

