Expression of CDK8 and CDK8-interacting Genes as Potential Biomarkers in Breast Cancer
- PMID: 26452386
- PMCID: PMC4755306
- DOI: 10.2174/156800961508151001105814
Expression of CDK8 and CDK8-interacting Genes as Potential Biomarkers in Breast Cancer
Abstract
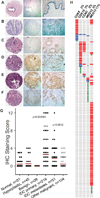
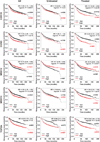
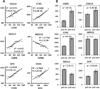
CDK8 and its paralog CDK19, in complex with CCNC, MED12 and MED13, are transcriptional regulators that mediate several carcinogenic pathways and the chemotherapy-induced tumor-supporting paracrine network. Following up on our previous observation that CDK8, CDK19 and CCNC RNA expression is associated with shorter relapse-free survival (RFS) in breast cancer, we now found by immunohistochemical analysis that CDK8/19 protein is overexpressed in invasive ductal carcinomas relative to non-malignant mammary tissues. Meta-analysis of transcriptomic data revealed that higher CDK8 expression is associated with shorter RFS in all molecular subtypes of breast cancer. These correlations were much stronger in patients who underwent systemic adjuvant therapy, suggesting that CDK8 impacts the failure of systemic therapy. The same associations were found for CDK19, CCNC and MED13. In contrast, MED12 showed the opposite association with a longer RFS. The expression levels of CDK8 in breast cancer samples were directly correlated with the expression of MYC, as well as CDK19, CCNC and MED13 but inversely correlated with MED12. CDK8, CDK19 and CCNC expression was strongly increased and MED12 expression was decreased in tumors with mutant p53. Gene amplification is the most frequent type of genetic alterations of CDK8, CDK19, CCNC and MED13 in breast cancers (9.7% of which have amplified MED13), whereas point mutations are more common in MED12. These results suggest that the expression of CDK8 and its interactive genes has a profound impact on the response to adjuvant therapy in breast cancer in accordance with the role of CDK8 in chemotherapy-induced tumor-supporting paracrine activities.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

