Parallelism and Epistasis in Skeletal Evolution Identified through Use of Phylogenomic Mapping Strategies
- PMID: 26452532
- PMCID: PMC5009992
- DOI: 10.1093/molbev/msv208
Parallelism and Epistasis in Skeletal Evolution Identified through Use of Phylogenomic Mapping Strategies
Abstract
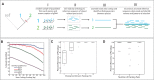
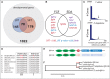
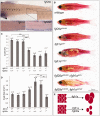
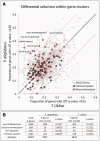
The identification of genetic mechanisms underlying evolutionary change is critical to our understanding of natural diversity, but is presently limited by the lack of genetic and genomic resources for most species. Here, we present a new comparative genomic approach that can be applied to a broad taxonomic sampling of nonmodel species to investigate the genetic basis of evolutionary change. Using our analysis pipeline, we show that duplication and divergence of fgfr1a is correlated with the reduction of scales within fishes of the genus Phoxinellus. As a parallel genetic mechanism is observed in scale-reduction within independent lineages of cypriniforms, our finding exposes significant developmental constraint guiding morphological evolution. In addition, we identified fixed variation in fgf20a within Phoxinellus and demonstrated that combinatorial loss-of-function of fgfr1a and fgf20a within zebrafish phenocopies the evolved scalation pattern. Together, these findings reveal epistatic interactions between fgfr1a and fgf20a as a developmental mechanism regulating skeletal variation among fishes.
Keywords: comparative genomics; epistasis; fgf signaling; nonmodel organisms; parallelism; zebrafish.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Altschul S, Gish W, Miller W. 1990. Basic Local Alignment Search Tool. J Mol Biol. 215:403–410. - PubMed
-
- Barrett RDH, Rogers SM, Schluter D. 2008. Natural selection on a major armor gene in threespine stickleback. Science 322:255–257. - PubMed
-
- Bell MA, Orti G, Walker JA, Koenings JP. 1993. Evolution of pelvic reduction in threespine stickleback fish: a test of competing hypotheses. Evolution 47:906–914. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

