SMIM1 is a type II transmembrane phosphoprotein and displays the Vel blood group antigen at its carboxyl-terminus
- PMID: 26452714
- PMCID: PMC4664579
- DOI: 10.1016/j.febslet.2015.09.029
SMIM1 is a type II transmembrane phosphoprotein and displays the Vel blood group antigen at its carboxyl-terminus
Abstract
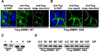
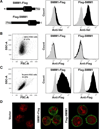
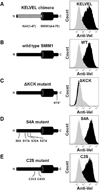
Disruption of SMIM1, encoding small integral membrane protein 1, is responsible for the Vel-negative blood type, a rare but clinically-important blood type. However, the exact nature of the Vel antigen and how it is presented by SMIM1 are poorly understood. Using mass spectrometry we found several sites of phosphorylation in the N-terminal region of SMIM1 and we found the initiating methionine of SMIM1 to be acetylated. Flow cytometry analyses of human erythroleukemia cells expressing N- or C-terminally Flag-tagged SMIM1, several point mutants of SMIM1, and a chimeric molecule between Kell and SMIM1 demonstrated that SMIM1 carries the Vel antigen as a type II membrane protein with a predicted C-terminal extracellular domain of only 3-12 amino acids.
Keywords: Blood group; Mass spectrometry; Phosphorylation; Small integral membrane protein; Type II membrane protein.
Copyright © 2015 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflicts of interest, financial or otherwise, regarding the publication of this manuscript.
Figures


References
-
- Storry JR, Joud M, Christophersen MK, Thuresson B, Akerstrom B, Sojka BN, Nilsson B, Olsson ML. Homozygosity for a null allele of SMIM1 defines the Vel-negative blood group phenotype. Nat Genet. 2013;45:537–541. - PubMed
-
- Sussman LN, Miller EB. New blood factor: Vel. Rev Hematol. 1952;7:368–371. - PubMed
-
- Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

