Functional and Structural Consequence of Rare Exonic Single Nucleotide Polymorphisms: One Story, Two Tales
- PMID: 26454016
- PMCID: PMC4684694
- DOI: 10.1093/gbe/evv191
Functional and Structural Consequence of Rare Exonic Single Nucleotide Polymorphisms: One Story, Two Tales
Abstract
Genetic variation arising from single nucleotide polymorphisms (SNPs) is ubiquitously found among human populations. While disease-causing variants are known in some cases, identifying functional or causative variants for most human diseases remains a challenging task. Rare SNPs, rather than common ones, are thought to be more important in the pathology of most human diseases. We propose that rare SNPs should be divided into two categories dependent on whether the minor alleles are derived or ancestral. Derived alleles are less likely to have been purified by evolutionary processes and may be more likely to induce deleterious effects. We therefore hypothesized that the rare SNPs with derived minor alleles would be more important for human diseases and predicted that these variants would have larger functional or structural consequences relative to the rare variants for which the minor alleles are ancestral. We systematically investigated the consequences of the exonic SNPs on protein function, mRNA structure, and translation. We found that the functional and structural consequences are more significant for the rare exonic variants for which the minor alleles are derived. However, this pattern is reversed when the minor alleles are ancestral. Thus, the rare exonic SNPs with derived minor alleles are more likely to be deleterious. Age estimation of rare SNPs confirms that these potentially deleterious SNPs are recently evolved in the human population. These results have important implications for understanding the function of genetic variations in human exonic regions and for prioritizing functional SNPs in genome-wide association studies of human diseases.
Keywords: RNA structure; ancestral allele; positive selection; purifying selection; single nucleotide polymorphisms; translational selection.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
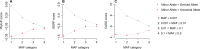


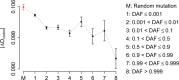

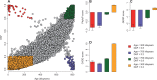
Similar articles
-
Evolutionary evidence of the effect of rare variants on disease etiology.Clin Genet. 2011 Mar;79(3):199-206. doi: 10.1111/j.1399-0004.2010.01535.x. Epub 2010 Sep 10. Clin Genet. 2011. PMID: 20831747 Free PMC article. Review.
-
Derived SNP alleles are used more frequently than ancestral alleles as risk-associated variants in common human diseases.J Bioinform Comput Biol. 2012 Apr;10(2):1241008. doi: 10.1142/S0219720012410089. J Bioinform Comput Biol. 2012. PMID: 22809343 Free PMC article.
-
Disease-associated alleles in genome-wide association studies are enriched for derived low frequency alleles relative to HapMap and neutral expectations.BMC Med Genomics. 2010 Dec 10;3:57. doi: 10.1186/1755-8794-3-57. BMC Med Genomics. 2010. PMID: 21143973 Free PMC article.
-
Genomic variations and distinct evolutionary rate of rare alleles in Arabidopsis thaliana.BMC Evol Biol. 2016 Jan 27;16:25. doi: 10.1186/s12862-016-0590-7. BMC Evol Biol. 2016. PMID: 26817829 Free PMC article.
-
An evolutionary framework for common diseases: the ancestral-susceptibility model.Trends Genet. 2005 Nov;21(11):596-601. doi: 10.1016/j.tig.2005.08.007. Epub 2005 Sep 8. Trends Genet. 2005. PMID: 16153740 Review.
Cited by
-
Analysis of pooled genome sequences from Djallonke and Sahelian sheep of Ghana reveals co-localisation of regions of reduced heterozygosity with candidate genes for disease resistance and adaptation to a tropical environment.BMC Genomics. 2019 Nov 7;20(1):816. doi: 10.1186/s12864-019-6198-8. BMC Genomics. 2019. PMID: 31699027 Free PMC article.
-
Genetic Susceptibility to Acute Kidney Injury.J Clin Med. 2021 Jul 8;10(14):3039. doi: 10.3390/jcm10143039. J Clin Med. 2021. PMID: 34300206 Free PMC article. Review.
-
Recent Updates on Corticosteroid-Induced Neuropsychiatric Disorders and Theranostic Advancements through Gene Editing Tools.Diagnostics (Basel). 2023 Jan 17;13(3):337. doi: 10.3390/diagnostics13030337. Diagnostics (Basel). 2023. PMID: 36766442 Free PMC article. Review.
-
Effects of Lipopolysaccharide-Binding Protein (LBP) Single Nucleotide Polymorphism (SNP) in Infections, Inflammatory Diseases, Metabolic Disorders and Cancers.Front Immunol. 2021 Jul 6;12:681810. doi: 10.3389/fimmu.2021.681810. eCollection 2021. Front Immunol. 2021. PMID: 34295331 Free PMC article. Review.
-
Temperature-Driven Intraspecific Diversity in Paralytic Shellfish Toxin Profiles of the Dinoflagellate Alexandrium pacificum and Intragenic Variation in the Saxitoxin Biosynthetic Gene, sxtA4.Microb Ecol. 2025 Aug 8;88(1):87. doi: 10.1007/s00248-025-02586-1. Microb Ecol. 2025. PMID: 40779244 Free PMC article.
References
-
- Argos P, Rao JK, Hargrave PA. 1982. Structural prediction of membrane-bound proteins. Eur J Biochem. 128:565–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

